Gogs
@gogulan_k
Co-founder & Head of Magnetic Resonance @bindresearch | Disordered proteins and Density matrices
ID: 829887799
17-09-2012 22:01:34
142 Tweet
171 Followers
449 Following

In the not too distant future I hope it's possible to do fully automated analysis of even the most complicated NMR experiments. I think #DeepLearning will be a big part of this. Here's our (D. Flemming Hansen) contribution to this effort. #NMRchat doi.org/10.26434/chemr…

I have a PhD position available in my group Oxford Biology to understand protein import mechanisms in chloroplasts using structural tools. Funded by @RoyalSociety and Oxford BBSRC DTP. The deadline is 7th January 2022. Contact me for more details plants.ox.ac.uk/dphil-projects… Please retweet!


Many thanks to Andy Byrd @NCIResearchCtr and his team for their Magnetic Resonance Letters highlight of our recent J. Am. Chem. Soc. paper lead by Gogs on #DeepLearning and #NMRchat UCL Faculty of Life Sciences Networks available at: github.com/gogulan-k/FID-… doi.org/10.1016/j.mrl.…





Need to drug an intrinsically disordered protein (#IDP)? Why not try 19F #NMR transverse spin-relaxation? Check out our most recent work, now on bioRxiv. D. Flemming Hansen vaibhav shukla Angelo M Figueiredo #NMRchat 🍝 💊 🧲 A 🧵👇🏽: (1/14) biorxiv.org/content/10.110…

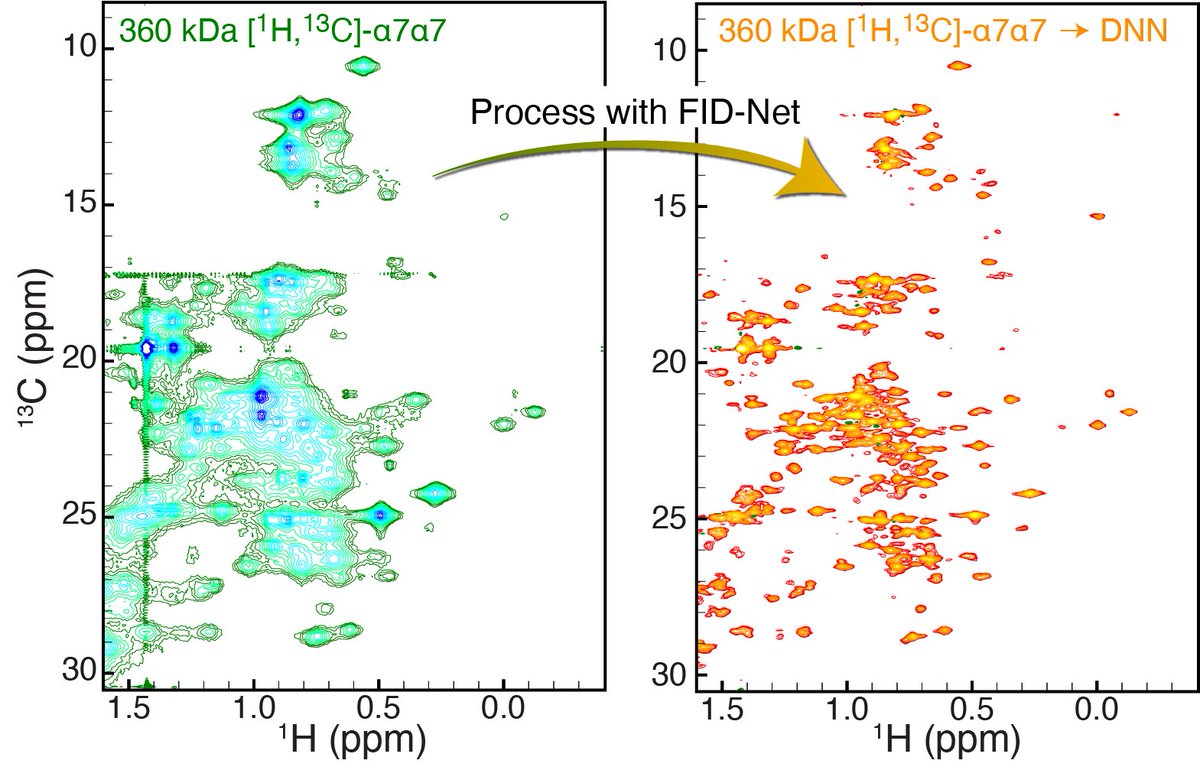
Our #NMRchat DNNs are now able to transform crowded HSQC spectra of large proteins (<350 kDa) into spectra akin to Methyl-TROSY (methyl-TROSY-fication). Read the full story on bioRxiv: doi.org/10.1101/2023.0… Engineering and Physical Sciences Research Council #DeepLearning UCL Faculty of Life Sciences Gogs vaibhav shukla

A bright future lies ahead for #NMR ! vaibhav shukla Gabi Heller and I have written a review about NMR in the era of AI. Maybe we are biased - but we think that a combination of AI and NMR will further revolutionise structural biology doi.org/10.1016/j.str.… UCL Faculty of Life Sciences


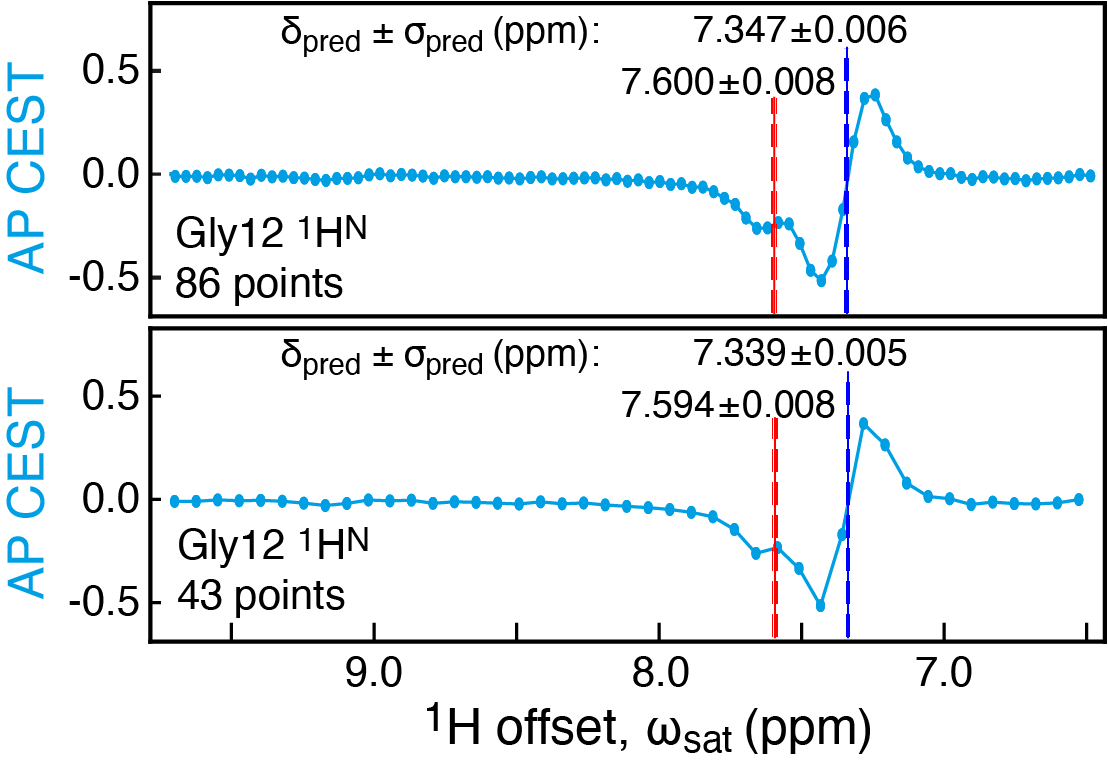
Happy to share FID-Net-2 to transform complex #NMR data👇 Using dedicated pulse sequences FID-Net-2 generates high-quality quantitative aromatic 13C-1H maps and accurate uncertainties for the transform. biorxiv.org/content/10.110… Gogs vaibhav shukla The Francis Crick Institute UCL Faculty of Life Sciences


Happy to share our DNNs to transform crowded methyl HSQC spectra of large proteins into spectra akin to Methyl-TROSY is out in Nature Communications Scripts & weights available on GitHub / Zenodo. #NMRChat Gogs vaibhav shukla UCL Faculty of Life Sciences The Francis Crick Institute rdcu.be/dKJGq


D. Flemming Hansen, Gogs and co-authors based The Francis Crick Institute and UCL Faculty of Life Sciences show deep neural networks can be trained to transform crowded NMR spectra of large proteins into readily interpretable spectra. #NMRChat, #DeepLearning nature.com/articles/s4146…


Delighted to be a part of this work demonstrating combined NMR pulse sequence design with deep neural network analysis to achieve high resolution spectra of aromatic side chains. D. Flemming Hansen vaibhav shukla Paper: science.org/doi/10.1126/sc… Code: github.com/gogulan-k/FID-…


I’m delighted to share that Thomas Löhr, Gogs, and I are launching Bind Research, a UK-based not-for-profit research startup to deliver publicly–available tools and datasets to make intrinsically disordered proteins druggable. 🍝 💊🧲 💻 🦠

Really excited to see Bind Research come to fruition! This is exactly what the IDP drug discovery field needs! Congrats to Gabi Heller, Thomas Löhr and Gogs! Hope to work with you all on this project soon!