Nate Gruver
@gruver_nate
Machine learning PhD student at NYU, BS & MS @StanfordAILab, Industry @AIatMeta @Waymo
ID: 1119059106231226370
https://ngruver.github.io/ 19-04-2019 02:04:16
128 Tweet
638 Followers
276 Following






LLMs are highly constrained biological sequence optimizers. In new work led by Angelica Chen & Samuel Stanton , we show how to drive an active learning loop for protein design with an LLM. 1/

I’m excited to share our latest work on generative models for materials called FlowLLM. FlowLLM combines Large Language Models and Riemannian Flow Matching in a simple, yet surprisingly effective way for generating materials. arxiv.org/abs/2410.23405 Benjamin Kurt Miller Ricky T. Q. Chen Brandon Wood

I am recruiting Ph.D. students for my new lab at New York University! Please apply, if you want to work with me on reasoning, reinforcement learning, understanding generalization and AI for science. Details on my website: izmailovpavel.github.io. Please spread the word!



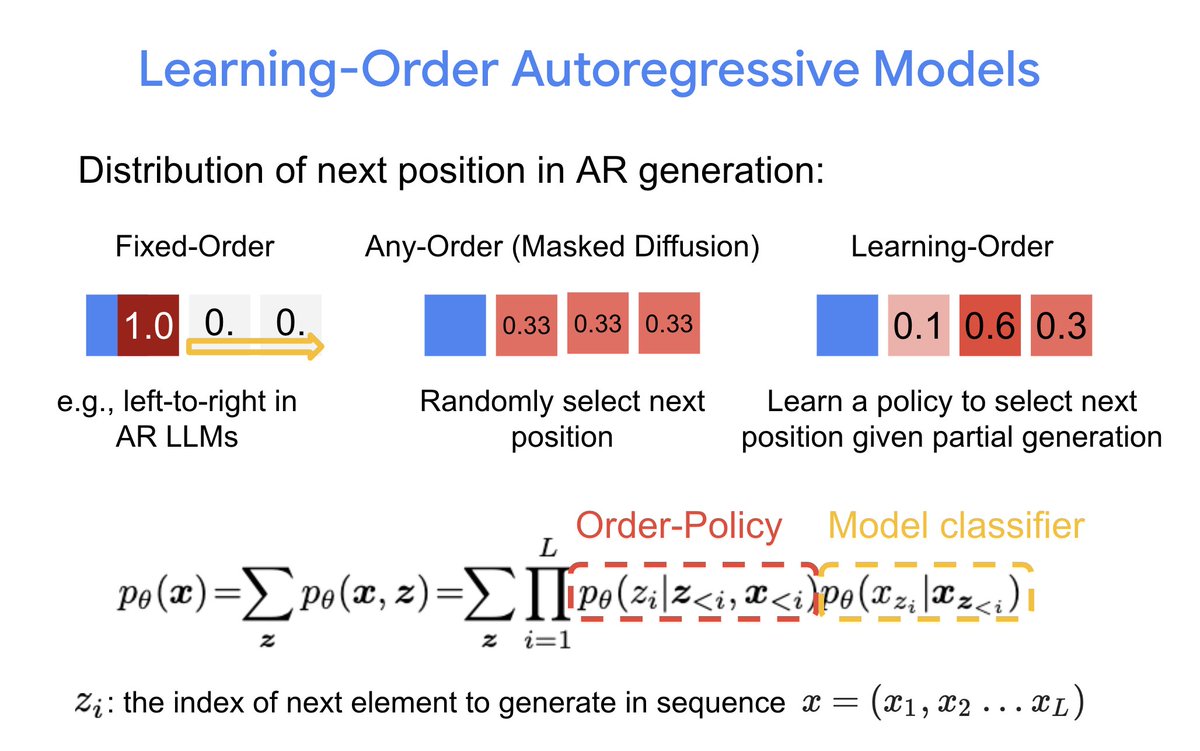
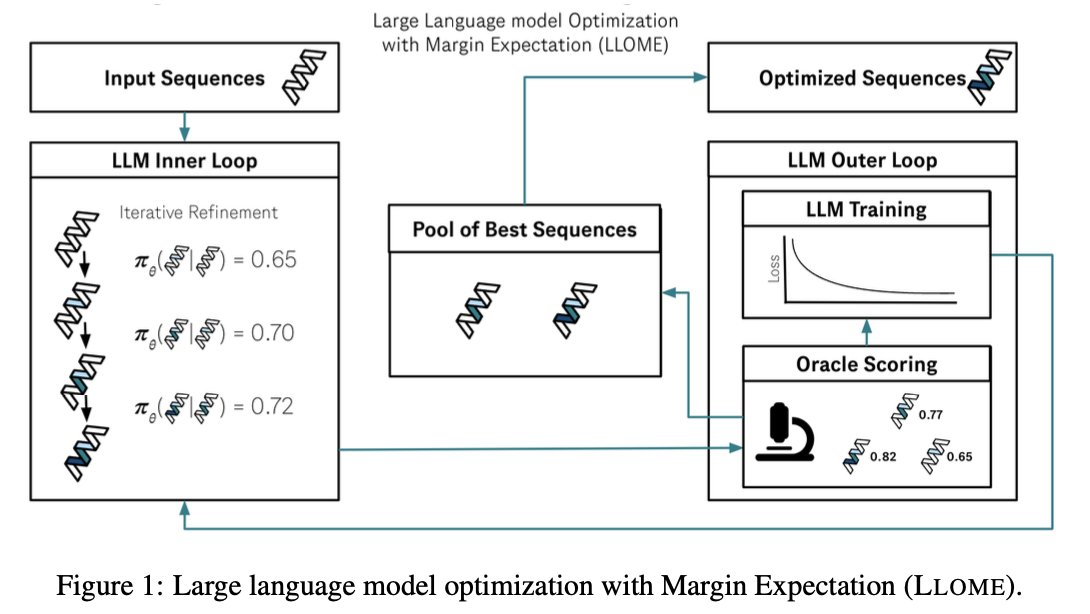
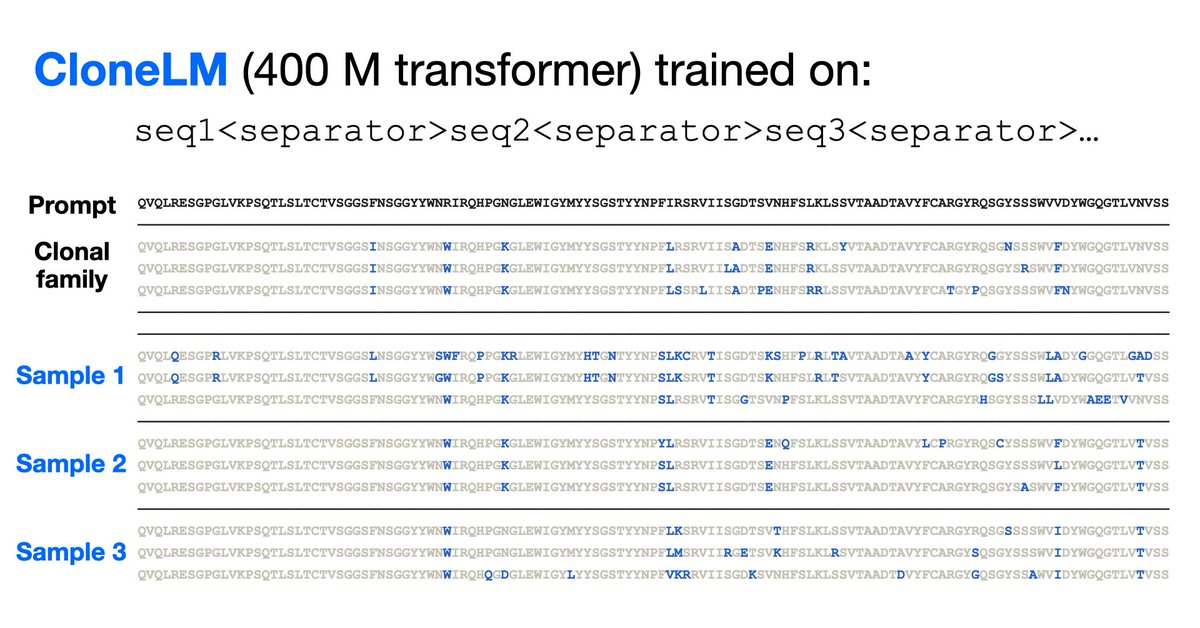
New model trained on new dataset of nearly a million evolving antibody families at AIDrugX workshop Sunday at 4:20 pm (#76) #Neurips! Collab between Andrew Gordon Wilson and BigHat Biosciences. Stay tuned for full thread on how we used the model to optimize antibodies in the lab in coming days!



Still more amazing professors signing on as potential coadvisors for our FutureHouse Fellowship. Kind of insane: -Debbie Marks (Harvard) Debora Marks -Brian Hie (Stanford/Arc) Brian Hie -Jonathan Gootenberg and Omar Abudayyeh (Jonathan Gootenberg Omar Abudayyeh) -Anima Anandkumar

To trust LLMs in deployment (e.g., agentic frameworks or for generating synthetic data), we should predict how well they will perform. Our paper shows that we can do this by simply asking black-box models multiple follow-up questions! w/ Marc Finzi and Zico Kolter 1/ 🧵