
ASHansen Lab
@hansen_lab
We seek to understand the molecular mechanisms that regulate genome organization and gene expression in time and space.
Based in @MITdeptofBE
ID: 1163878481727377413
https://www.ashansenlab.com/ 20-08-2019 18:20:43
82 Tweet
930 Followers
18 Following


Excited to welcome Jack Toppen to the lab as a new graduate student who joins us from the MIT CSB PhD Program graduate program - Jack is interested in developing computational tools for understanding and modeling transcriptional bursting.



We are thrilled to welcome Coco Wu to the lab as a new graduate student from the MIT Biology graduate program co-advised with Francisco J. Sánchez-Rivera - Coco is interested in exploring the relationship between DNA sequence, DNA looping, gene expression, and disease!




Excited to welcome Jacob Jacob Kæstel-Hansen as a new Novo Nordisk Foundation post-doc to the lab!




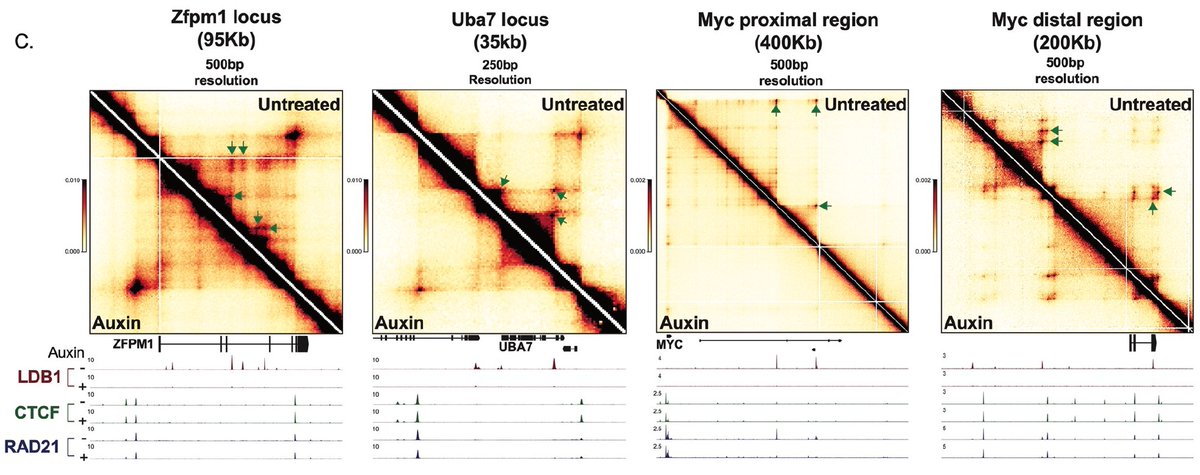
Congrats to Nicholas Aboreden blobellab with RCMC contribution from Viraat Goel ASHansen Lab . Having recently worked on what we thought was a looping factor (ZNF143 biorxiv.org/content/10.110… ) it was amazing to work on a strong Enh-Promoter looping factor LDB1 biorxiv.org/content/10.110…

Congrats to Gavin Schlissel Pulin Li with single-molecule imaging contribution from Domenic Narducci ASHansen Lab Single-molecule imaging of hedgehog in morphogen gradients reveals a surprisingly complex diffusion mechanism: pnas.org/doi/10.1073/pn…


Congrats to Viraat Goel James Jusuf et al. on new preprint in collab with @Irate_Physicist blobellab Leonid Mirny discovering transient microcompartments that connect enhancers and promoters in mitotic chromosomes: biorxiv.org/content/10.110…

Very grateful to NIH Common Fund for funding our Transformative Research Award #NIHHighRisk


Our preprint by Domenic Narducci and the companion preprint by deWitLab are now out in Molecular Cell ZNF143 is not a general looping factor and was identified as such due to a non-specific antibody. Instead it is TF with the longest residence time measured (to our knowledge)





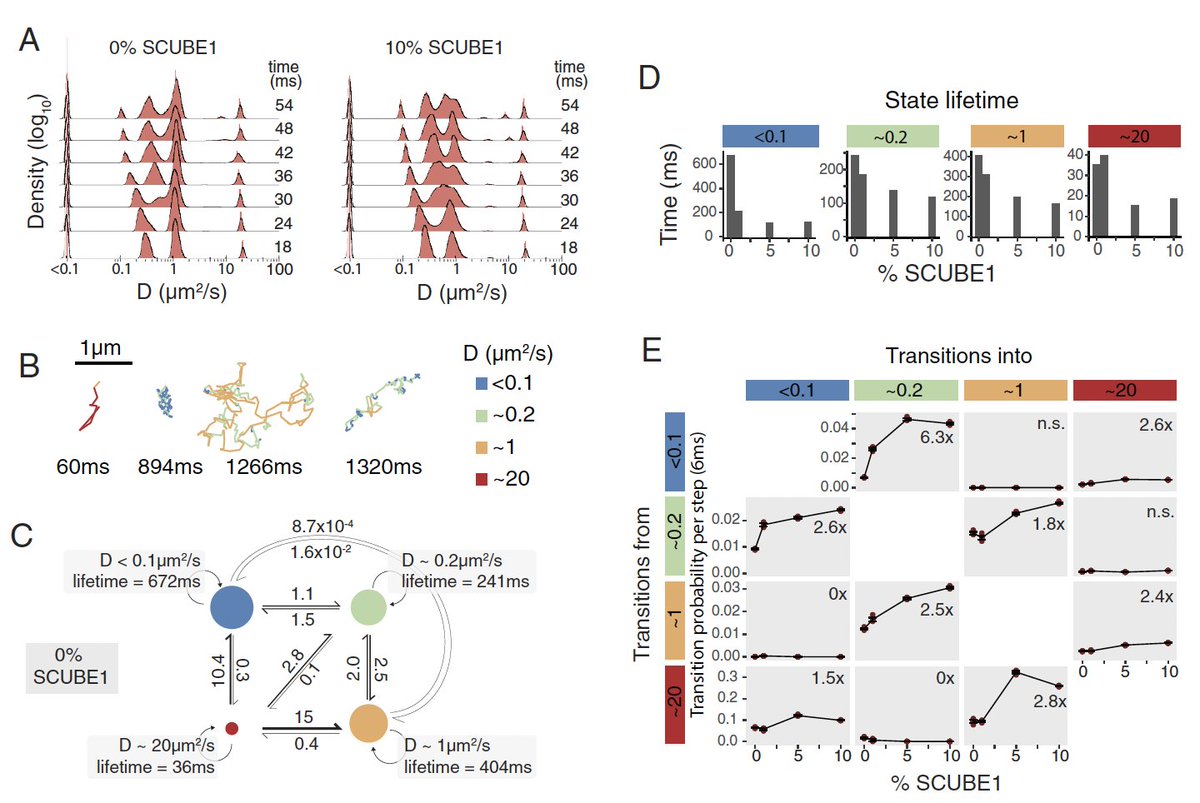
(1/13) Thread on Matteo Mazzocca Domenic Narducci Simon Grosse-Holz Jessica Matthias new preprint Q: how does chromatin move? Using MINFLUX, SPT & SRLCI, we track chromatin dynamics across 7 orders of magnitude in time to provide some answers biorxiv.org/content/10.110…









