Hao Lawrence Huang
@haohuang1999
ID: 1458637035523100674
11-11-2021 03:27:20
23 Tweet
23 Followers
57 Following





We held a reading group on Transformers (watched videos / read blog posts / studied papers by Lucas Beyer (bl16) Andrej Karpathy Chris Olah Aman Arora Jay Alammar Sasha Rush et al.), and now I _finally_ roughly understand what attention does. Here is my take on it. A summary thread. 1/n



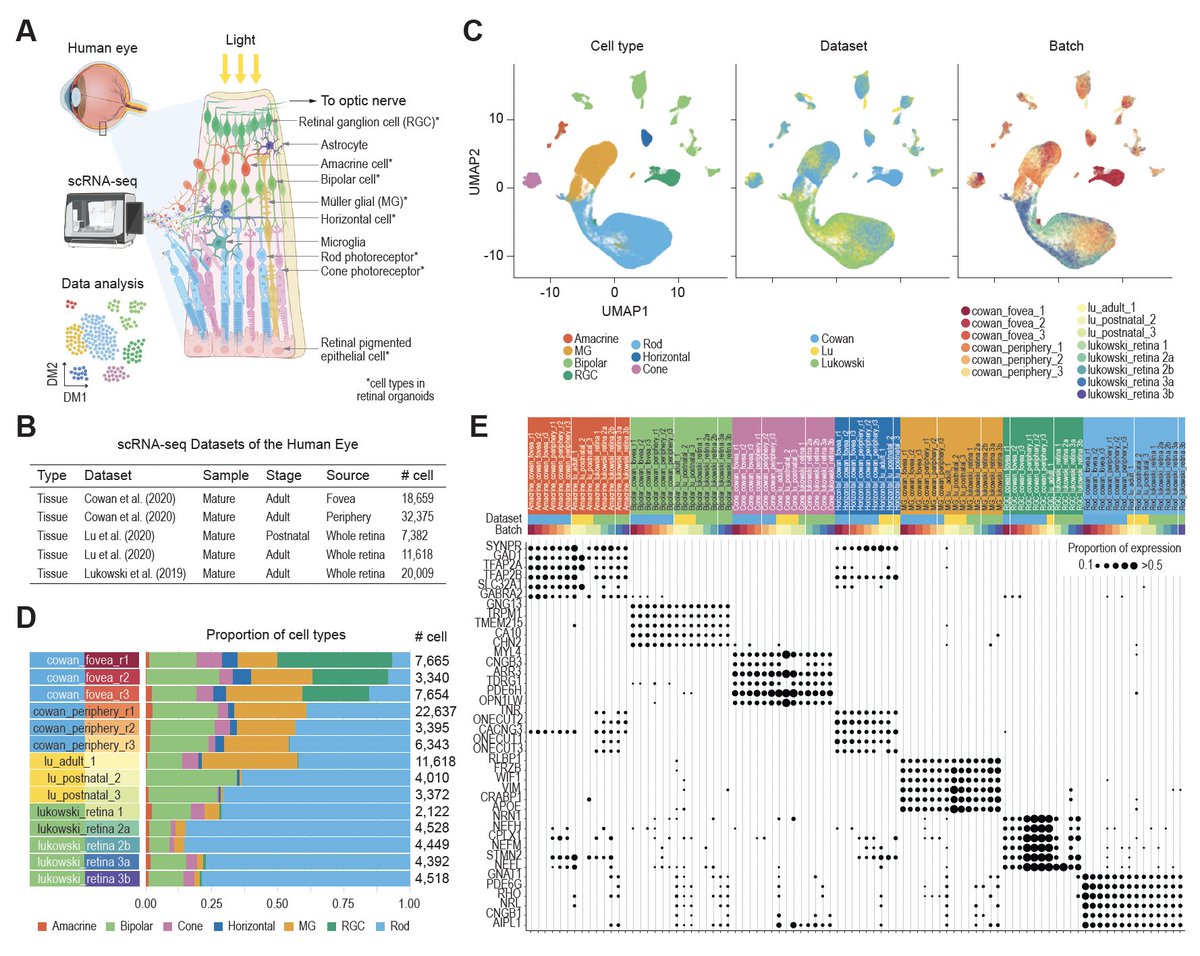
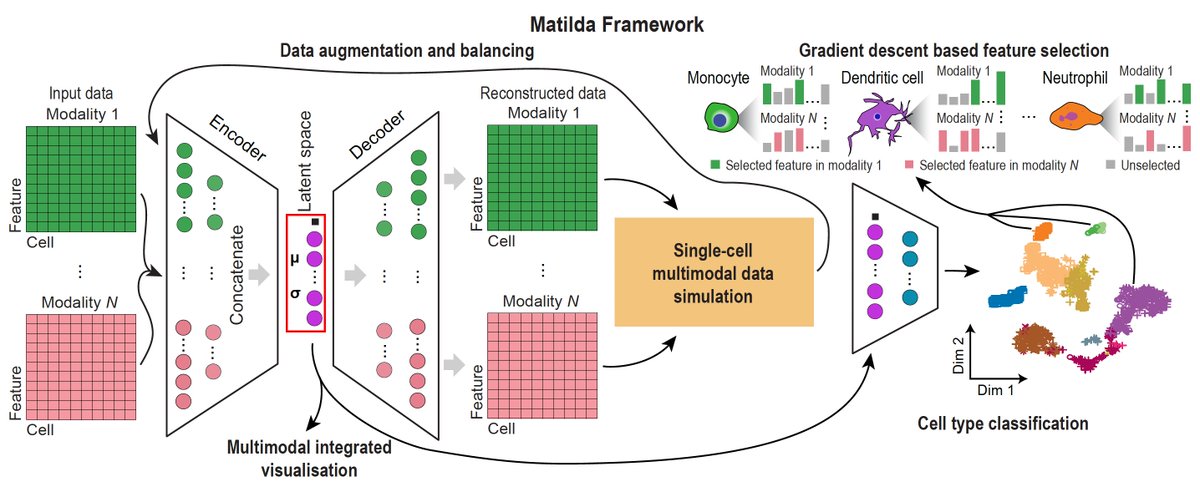
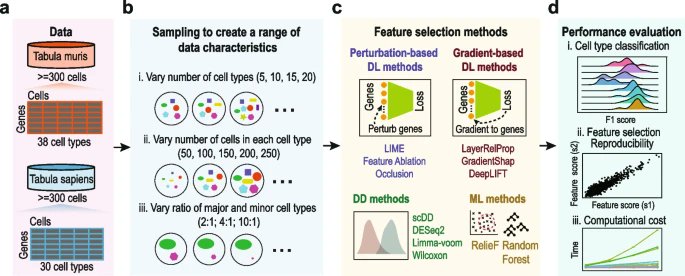
New research from Computational Systems Biology Lab Hao Lawrence Huang & co. University of Sydney providing a reference for future development and application of #deeplearning based feature selection methods for single-cell omics data analyses bit.ly/47xz54N

Demuxafy: improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods. url.au.m.mimecastprotect.com/s/dmyICmO5nJhp… Led by Drew Neavin in collab with the sc-eQTLGen Consortium Lude Franke Shyam Prabhakar Lab Jimmie Ye @davisjmcc Marta Melé Martin Hemberg

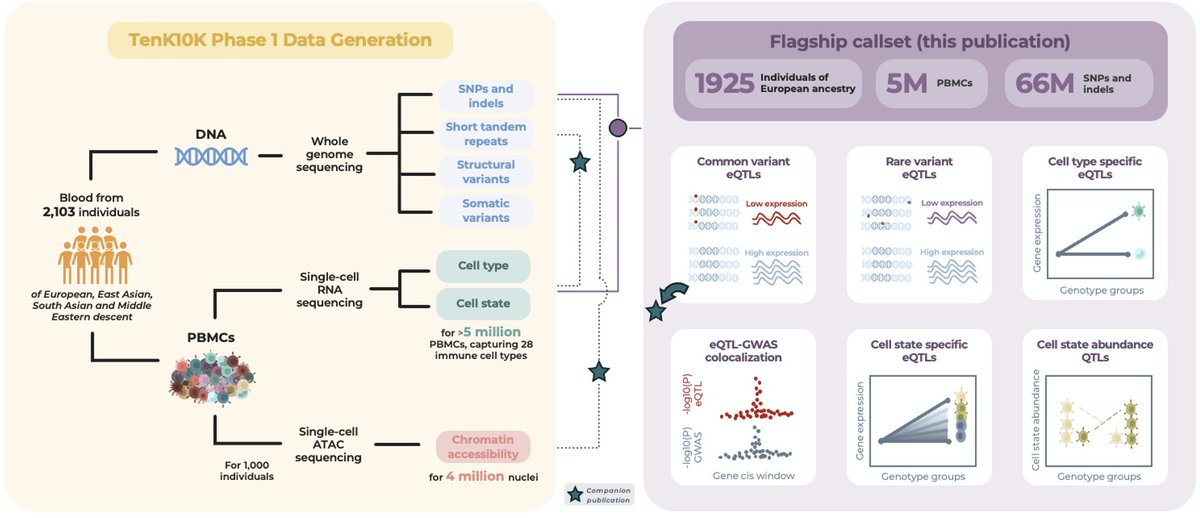
We are thrilled to announce the official launch of Joseph Powell's newest project, TenK10K. Thank you to the Hon. David Harris MP, Simon Giuliano, Illumina and Benjamin Kile, Garvan Institute of Medical Research for officially launching TenK10K! @cellularfutures UNSW Medicine & Health Ramaciotti Centre for Genomics


I am thrilled to have been awarded NHMRC Investigator Grant (EL1). My research will be focused on understanding the genetic regulation of autoimmune diseases at the cellular level by building a population-scale single-cell atlas of immune cells. Garvan Institute of Medical Research UNSW Medicine & Health

Garvan researchers Dr Drew Neavin and Prof Joseph Powell have been awarded a prestigious Data Insights grant from Chan Zuckerberg Initiative. The funding will support their groundbreaking 18-month project, SNAPSHOT, aimed at understanding cellular variation in health and disease.