
Tazro Inutano Ohta
@inutano
#WeAreHackersWeDance
ID: 198902933
http://github.com/inutano 05-10-2010 14:58:56
1,1K Tweet
361 Followers
292 Following

Watch Tazro Ohta (Tazro Inutano Ohta) talk about sharing reusable computational workflows with his new Yevis platform in our youtube channel youtu.be/Yr7UUw_lqGE


A workflow reproducibility scale for automatic validation of biological interpretation results Tazro Inutano Ohta et al propose a new metric, a reproducibility scale of workflow execution results, to evaluate the reproducibility of computational results. doi.org/10.1093/gigasc…
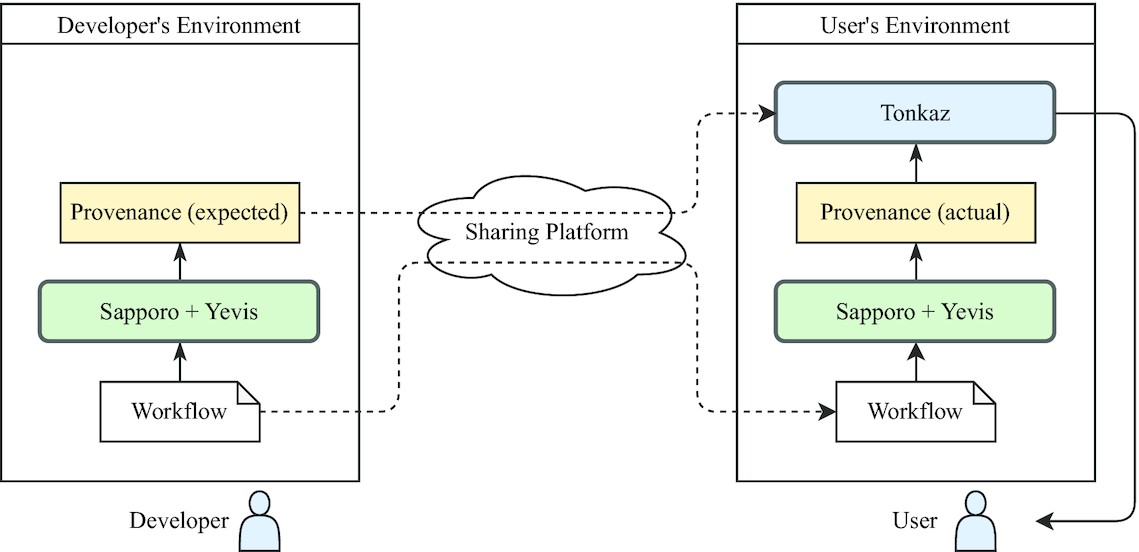

This workflow utilises their recently published Yevis workflow registry, and Tazro Inutano Ohta talks more about this in a recent video youtu.be/Yr7UUw_lqGE



Thanks to everybody who made #biohackjp so exceptional and to BioHackathon toshiaki katayama for the invitation. Hopefully we meet again this autumn for #BioHackEU23




BioThings API toolsets for building & integrating biomedical knowledge #API at #bosc2023 #ISMBECCB2023. Visit our posters today and/or join two talks on BioThings SDK and BioThings Explorer (by Jackson Callaghan) tomorrow, day-2 of #bosc2023


Was great to see Tazro Inutano Ohta presenting in #BOSC2023 on Tonkaz, his recent paper in GigaScience presenting a workflow reproducibility scale for automatic validation of biological interpretation results doi.org/10.1093/gigasc… #ISMBECCB2023



In addition, a new tool, Diff Analysis, has been released that allows the retrieval of differentially methylated regions (DMRs) from two given sets of WGBS data. Thanks to Tazro Inutano Ohta and ZOU, Zhaonan (鄒 兆南) for their great contributions!




Good morning Japan! Mount Fuji looking amazing with fresh snow. I have a week talking to Japanese colleagues Bioinformation and DDBJ Center DNA and metabolomics data sharing for the benefit of the world!


#ChIPAtlas Our paper about a major update of ChIP-Atlas is out! ChIP-Atlas 3.0 provides not only ChIP-seq, ATAC-seq and Bisulfite-seq data, but also Hi-C, ChromHMM, GWAS SNPs and eQTLs data. Great contribution from ZOU, Zhaonan (鄒 兆南) and Tazro Inutano Ohta ! academic.oup.com/nar/article/do…





