
Jakob Woerner
@jakob_woerner
Genomics and Computational Biology PhD Student at UPenn 🧬
ID: 1407422039636557829
22-06-2021 19:35:56
18 Tweet
129 Followers
236 Following


Glad to share our story: “Circulating microbial content in myeloid malignancy patients is associated with disease subtypes and patient outcomes” out in Nature Communications! Always a nice team effort!! 👉 doi.org/10.1038/s41467…


Another great day from the Kim lab! These young scientists have been inspiring me everyday!! Anni Moore Jakob Woerner did a fantastic job during the 2nd rotation. Welcome, K S Manu & Matthew Lee for your 3rd rotation. UPenn GCB Penn IBI



I will be a conference chair of #MABC2022 (Oct 7). We have a list of excellent speakers. The theme is "Computational Approaches to Differentiation in Diseases." Submit your abstract due Aug 15. #bioinformatics CHOP Department of Biomedical & Health Informatics Penn IBI Penn Medicine UPenn GCB midatlanticbioinformatics.org




Looking forward to sharing some of our most recent work today at #ASHG2022 in collaboration with Jakob Woerner, Yonghyun Nam, and Dokyoon Kim! Come check us out at PB3265 from 3 - 4:45 PM #Genomics #NetworkScience #NetworkMedicine


Team Artificial Intelligence now shows its project SynergAIze - #AI applied in cancer treatment focusing on kinases. 👍 Emilio Dorigatti Giulia Cantini @[email protected] Jakob Woerner Kenneth Verstraete Pauline L. Pfuderer Julia Kacher #science #InnoCup23 #research #phdstudent merckgroup.com/en/research/op…


Team #AI (project SynergAIze) is 1st runner up in today's #InnoCup23 - congratulations! Emilio Dorigatti Giulia Cantini @[email protected] Jakob Woerner Kenneth Verstraete Pauline L. Pfuderer Julia Kacher Karin Hrovatin Marsico Lab @[email protected] CRUK Cambridge Centre Universität München #science #innovation merckgroup.com/en/research/op…


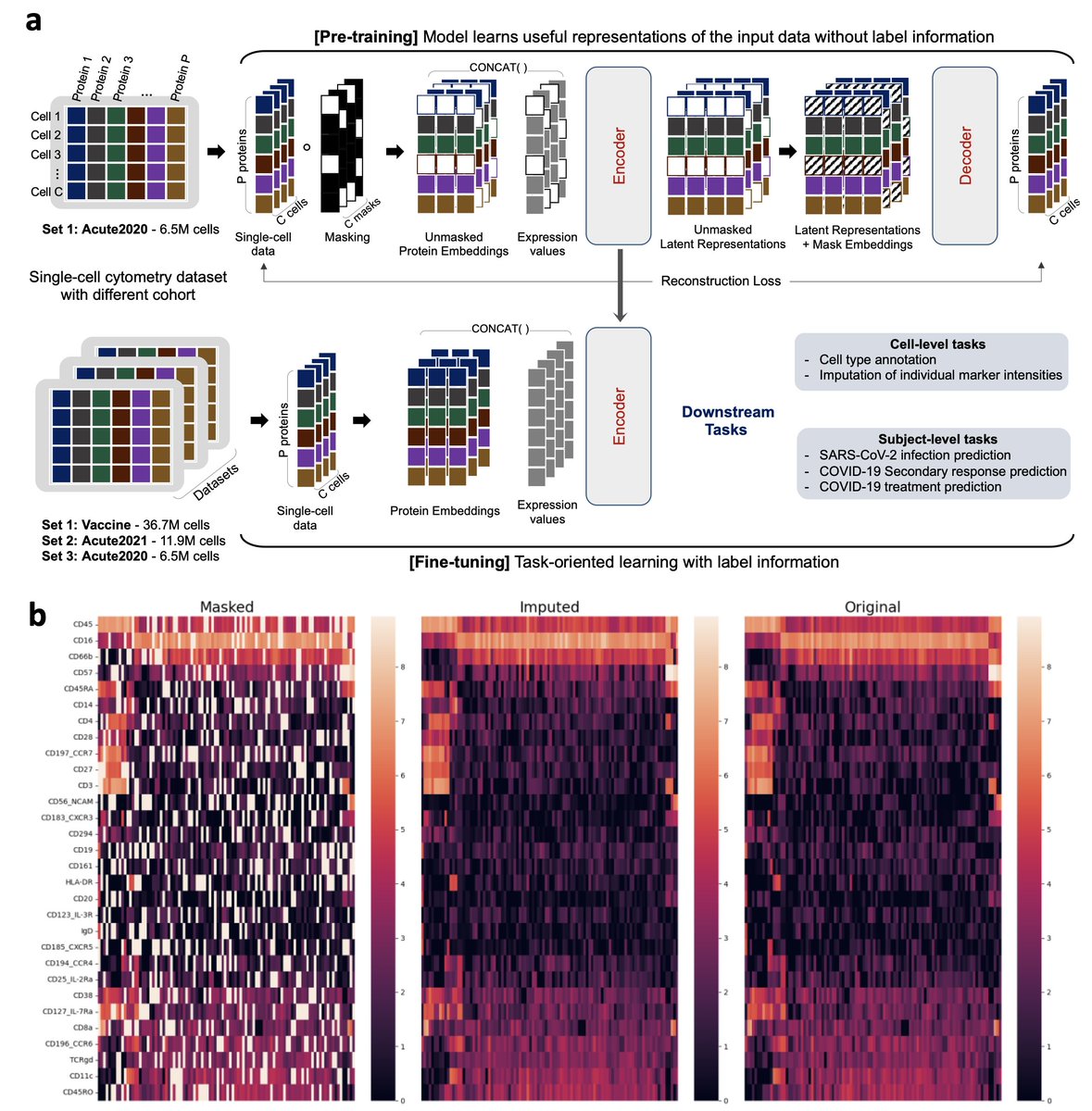
Excited to share our recent work: "#SingleCell Masked Autoencoder: An Accurate and Interpretable Automated Immunophenotyper" scMAE model transforms high-throughput single-cell cytometry, offering a scalable, robust, and precise immunophenotyping tool.E. John Wherry Immune Health

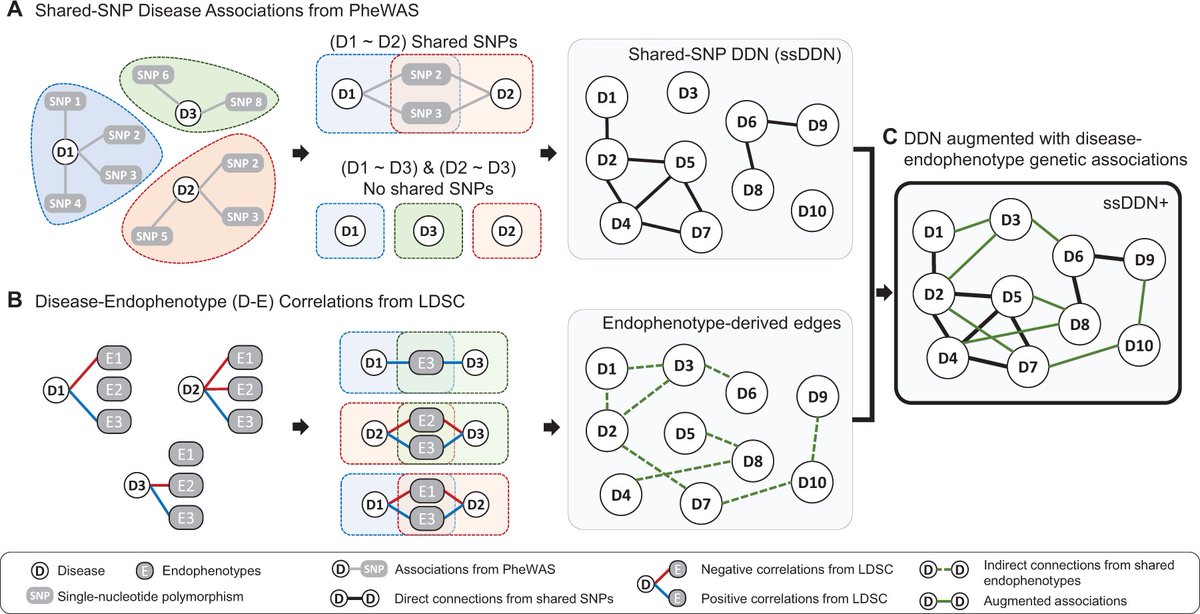
Excited that our work investigating the value of quantitative endophenotypes in disease network analysis is now out in Bioinformatics! Very exciting collaboration with my two favorite mentors Vivek Sriram and Dokyoon Kim doi.org/10.1093/bioinf…

New research identifies genetic associations in the human diseasome using an endophenotype-augmented disease network ft. Jakob Woerner, Vivek Sriram (UPenn GCB), Anurag Verma (Penn Medicine), Yonghyun Nam & Dokyoon Kim (Dept. of Biostatistics, Epidemiology & Informatics) Penn IBI tinyurl.com/bdebj2c8


Our group has contributed to three platform talks and four posters at #ASHG24 this year applying #HPC and #AI to Human Genetics! I am proud of all we were able to accomplish this year. cc Anurag Verma Hae Kyung Im 任慧耕 임혜경


Excited to present at #ASHG24 this week on our work analyzing human phenotype networks using a meta-analysis of 1.8 million individuals. You can hear my talk at the "Rare Variants and Admixture Modeling in Diverse Populations" session on Friday at 2! Anurag Verma Dokyoon Kim

🚨 Don’t miss this! My grad student Jakob Woerner will present: "Network comparison of ancestry-specific genetically correlated diseases in a meta-analysis of #PheWAS from 1M individuals" 🗓️ Nov 8 | ⏰ 2:00 PM | 📍 Four Seasons Ballroom 2&3 #ASHG24 UPenn GCB Anurag Verma

Very excited to share our preprint led by Michael Levin Satoshi Koyama Jakob Woerner & with Scott Damrauer assessing genome-wide pleiotropy of >1,000 clinical traits across ~1.7M individuals with nearly 30K locus-trait associations! medrxiv.org/content/10.110… medRxiv [1/3]
![Pradeep Natarajan (@pnatarajanmd) on Twitter photo Very excited to share our preprint led by <a href="/MGLevin/">Michael Levin</a> <a href="/skoyamamd/">Satoshi Koyama</a> <a href="/jakob_woerner/">Jakob Woerner</a> & with <a href="/damrauer/">Scott Damrauer</a> assessing genome-wide pleiotropy of >1,000 clinical traits across ~1.7M individuals with nearly 30K locus-trait associations!
medrxiv.org/content/10.110… <a href="/medrxivpreprint/">medRxiv</a>
[1/3] Very excited to share our preprint led by <a href="/MGLevin/">Michael Levin</a> <a href="/skoyamamd/">Satoshi Koyama</a> <a href="/jakob_woerner/">Jakob Woerner</a> & with <a href="/damrauer/">Scott Damrauer</a> assessing genome-wide pleiotropy of >1,000 clinical traits across ~1.7M individuals with nearly 30K locus-trait associations!
medrxiv.org/content/10.110… <a href="/medrxivpreprint/">medRxiv</a>
[1/3]](https://pbs.twimg.com/media/GpN2beJWMAAyF55.jpg)

