
JASPAR database
@jaspar_db
JASPAR is an #openaccess database of manually curated, non-redundant transcription factor (TF) binding profiles
ID: 919854537132912640
http://jaspar.genereg.net/ 16-10-2017 09:16:26
83 Tweet
330 Followers
53 Following


Native JASPAR database transcription factor tracks are now available for hg19 and hg38. JASPAR has long been a popular public hub, and that hub will continue to be accessible and offers annotations on additional assemblies. See our news for more: bit.ly/UCSCjaspar

Happy to share that we have added eight excellent Norwegian bioinformatics services to our service delivery plan with ELIXIR Europe! Read more here👇 elixir.no/news/74/63/Eig…

Excited that JASPAR 2022 is out today! It's a significant update, with new TF binding profiles, new enrichment analysis tools and new word cloud profiles. Have at it everyone. #JASPAR2022 bit.ly/3o5dU6k Great JASPAR database team effort catalyzed by Anthony Mathelier

Great to share our new manuscript describing the latest update of JASPAR JASPAR database. New TF binding profiles, familial binding profiles, structural classification of plant DBDs, genome tracks, TFBS enrichment tool, and a new Python package, pyJASPAR. academic.oup.com/nar/advance-ar…

We are happy to announce the release of JASPAR Transcription Factor Binding Sites for hg19, hg38, mm10, and mm39. This release corresponds to JASPAR CORE 2022. Learn more about this release at genome.ucsc.edu/goldenPath/new… JASPAR database Anthony Mathelier

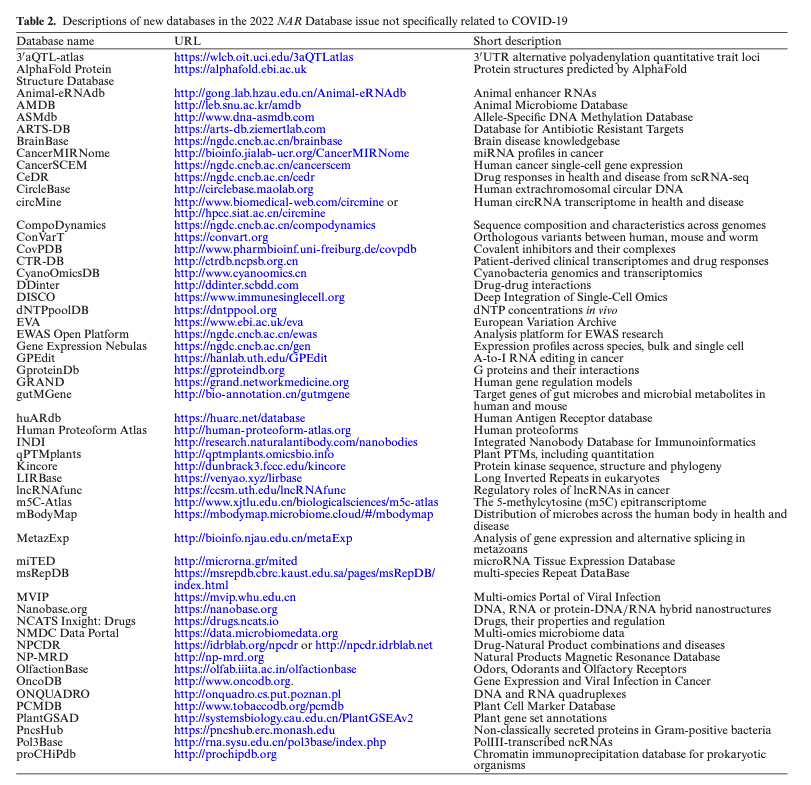
The Nucleic Acids Res Database Issue 2022 is online with 87 new databases and 85 updates, including JASPAR database academic.oup.com/nar/issue/50/D1 Here is the complete list of new databases with URLs - academic.oup.com/view-large/325…

JASPAR database The problem seems to be fixed. JASPAR is back up and running! JASPAR database #JASPAR

After 18 years, the original JASPAR_DB paper has reached 2000 citations. Glad it has been useful. doi.org/10.1093/nar/gk… The all-star team: @[email protected] Wynand Alkema par_engstrom BorisLenhard

Great day with this amazing group of people! It was such an enjoyable full day of JASPAR JASPAR database hackathon in sunny Oslo. Really lovely to change the scenery for pushing the next JASPAR release and get together. I'm so lucky to work with such fantastic people!!!


@Francois_Parcy JM Franco Zorrilla TrendsPlantSci Romain Blanc-Mathieu Awesome work! Congrats Romain Blanc-Mathieu @Francois_Parcy et al. Very valuable for the community; and for the JASPAR JASPAR database classification 😁😉

News article on our most recent publication 👇. Thanks @NCMMnews, and of course Tatiana Belova for the great work ✨ med.uio.no/ncmm/english/n…


#JobVacancy ELIXIR Norway, location Informatikk på UiB, is hiring System #developers and System #administrators to work on our exciting #Bioinformatics #infrastructure. jobbnorge.no/en/available-j… jobbnorge.no/en/available-j…



🎉 Excited to announce the 10th release & 20th-anniversary update of JASPAR JASPAR database , a leading resource in computational regulatory genomics! Important to note that JASPAR has a new URL: jaspar.elixir.no The manuscript can be found at academic.oup.com/nar/advance-ar…. 1/9

We are excited to announce the new JASPAR 2024 tracks for hg19, hg38, mm10, and mm39 which represent genome-wide predicted binding sites for transcription factors with binding profiles in the JASPAR CORE collection (JASPAR database). Learn more at bit.ly/JASPAR2024

I am very excited to see the outcome of the extensive work provided by ELIXIR Norway Oslo to support JASPAR JASPAR database in its deployment, maintenance, and monitoring, during the ELIXIR All Hands meeting. Great work Jeanne Chèneby, Sveinung Gundersen, Morten Johansen, and Eivind Hovig.
