Fengbin Wang (Jerry)
@jerrynosnothing
Principal Investigator @UAB_Biochem, protein/peptide scientist working on microbial pili and nanotube design | Views are my own | 😺⚡️👨🔬🍷☕️
ID: 1316923966141988865
https://jerryuab.org 16-10-2020 02:08:57
173 Tweet
455 Followers
328 Following


Excited to share the first paper from our lab is out as a preprint! Joshua Huffines established a dual-species culture model to investigate S. aureus-Corynebacterium interactions on nasal cells & how temperature influences bacterial persistence & cytotoxicity

The #cryoEM research centers UW–Madison Biochem are recruiting! We seek to hire an #administrative #assistant to join our team. We are looking for someone who has a passion for science, organization, communications, and outreach. Take a look! DM & RT! jobs.wisc.edu/jobs/administr…





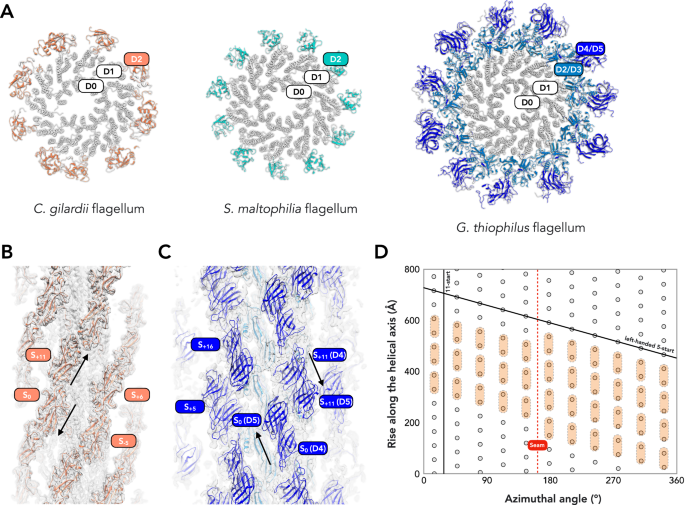
Check out our new cryo-EM preprint on bacterial flagellar outer domain diversity! No clue who'll see this due to Twitter's new algorithm. Important points: most work was done by rotation students, and collaborating with the Wu lab is great. HWU-OHSUSoD 🦠biorxiv.org/content/10.110…

I guess our research future is bright... Vincent Conticello I didn't see this coming #3BodyProblem 🍷👽


🔬👩🔬 Exciting news! I'll soon start my lab Van Andel Institute (VAI) to research molecular mechanisms in Neurodegenerative diseases. 🧠💡 Now recruiting postdocs to join our dynamic research team! Email me if you're interested! #Postdoc #CryoEM #Neuroscience #ResearchOpportunity



🚨 We've updated the glycan structure database on GlycoShape (glycoshape.org)! You can now search through and rebuild glycoproteins with 453 unique glycans 🥳 Glycans are now primarily listed by their GlyTouCan IDs Gly Kiyoko Kinoshita 🎉 #glycotime


Thank you Fengbin (Jerry) Wang Fengbin Wang (Jerry) for inviting me to speak at UAB_Biochem and meet Gino Cingolani Cingolani Lab, Laura Volpicelli-Daley Laura Volpicelli, Lindsay Rizzardi Lindsay Rizzardi, PhD, Peter Prevelige, Matthew Renfrow, Shu Chen, Louise Chow, and Tom Broker.


Researchers led by Phil Shuaiqi Guo, Jun Liu Lab, Lori L. Burrows report the structure of the Pseudomonas aeruginosa T4P suggesting the localization and role of its tip adhesin, PilY1. nature.com/articles/s4146…


Fengbin Wang (Jerry) @mkrupovic HWU-OHSUSoD used cryo-EM to determine the structures of three bacterial flagellar filaments, showing distinct outer domains. nature.com/articles/s4146…


1/ Thrilled to announce Dr. Elizabeth Wright from UW–Madison Biochem as the inaugural winner of the June Almeida Award for #cryoem! 🎉Elizabeth’s pioneering work in both cryo-ET methods and structural virology made her a perfect fit for this award. Congrats, Elizabeth Wright! ❄️🔬

Donor Strand Complementation and Calcium Ion Coordination Drive the Chaperone-free Polymerization of Archaeal Cannulae biorxiv.org/content/10.110… Happy that the preprint is now published. Great collaboration with fantastic colleagues. conticellolab Emory Chemistry

Another wonderful collaboration with Alan Brown lab. The Zhang lab together with Polina Lishko lab (co-first author Qingwei Niu) contribute the doublet microtubule structure from Crithidia fasciculata, a model organism that infects mosquitoes. science.org/doi/10.1126/sc…