
Joseph Replogle
@josephmreplogle
MD PhD, PGY1 at @MGHMedicine/@HarvardMed
ID: 1448811011314757632
15-10-2021 00:46:14
41 Tweet
729 Followers
195 Following

Fun to see the Perturb-seq toolbox continue to expand from Rahul Satija Neville Sanjana. Cas13 Perturb-seq has been on the tech with list! Capture of barcode gRNA at 3' end of tandem Cas13 guide array = less sgrna recombination and higher lenti titers.


Our latest CRISPRi screening tools + tips w/ @JSWlab and Marco Jost: (1) dual-sgRNA libraries ⬆ knockdown while decreasing library size (2) comparison of CRISPRi effectors finds that Zim3-dCas9 offers ⬆ knockdown with minimal toxicity (3) validated Zim3-dCas9 cell lines



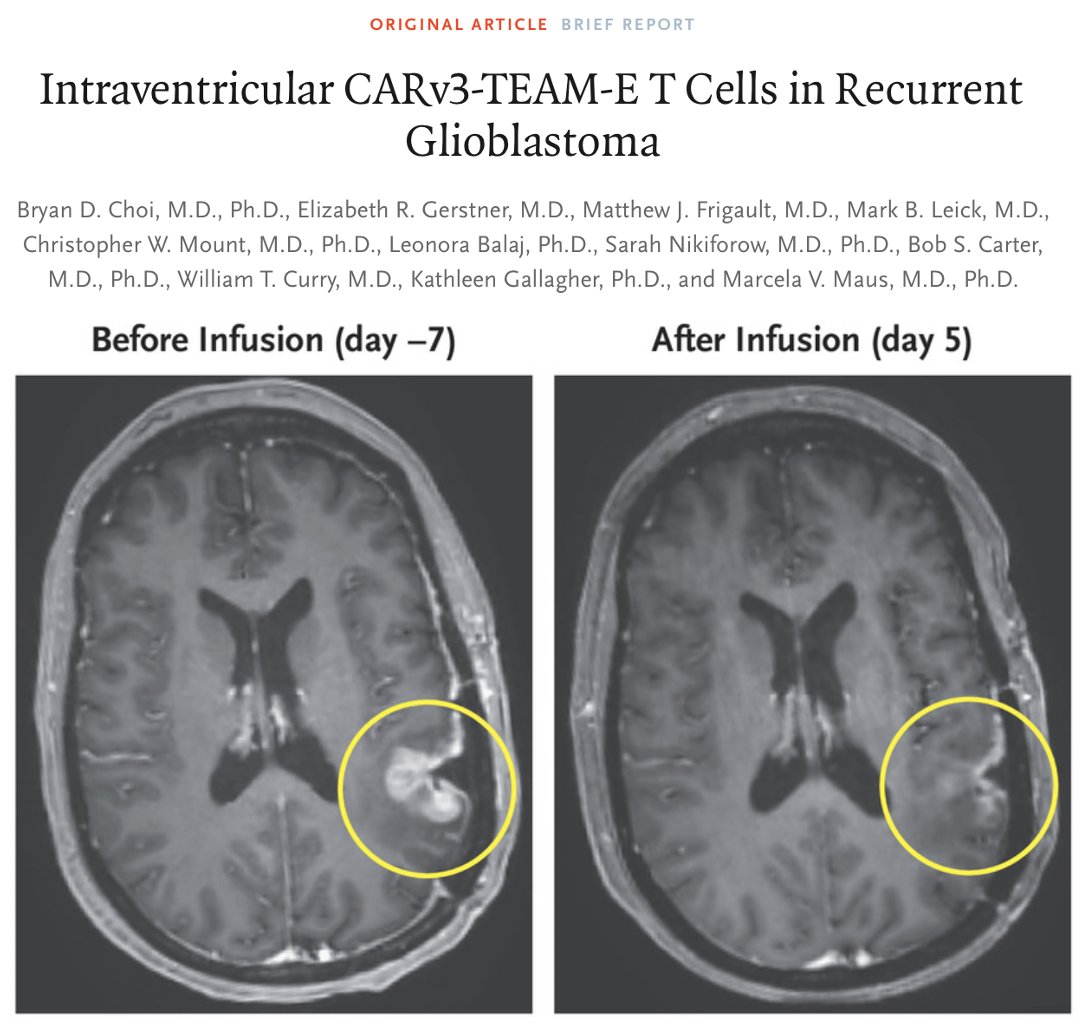
Today, NEJM published our clinical study of a novel #CARTCell in patients with #Glioblastoma. Early results with CARv3-TEAM-E suggest safety and bioactivity. Special thanks to all collaborators MGH Neurosurgery and Mass General Cancer Center! nej.md/3VnjUIq

From Russell Walton Blainey Lab , a very useful vector for those interested in taking advantage of sgRNA multiplexing for single cell CRISPR screens!


Check out our latest large-scale Perturb-seq on biorxiv! biorxiv.org/content/10.110… A new stat gen/GWAS-inspired analytic framework + large-scale screens in an expanded range of cell types Thanks Ajay Nadig and Luke O'Connor for involving us!


Just how impactful are large-scale perturbational data in single cell genomics? In the last two weeks, there have been *three* modeling/stats preprints with qualitatively different questions and insights for the same dataset generated by Joseph Replogle Jonathan Weissman's Lab (links below)




Even while in residency and with infants at home, I find it hard not to put everything down to read this new analysis of our data by Jonathan Pritchard and team! And in the same week others are working toward optimizing large-scale Perturb-seq in cardiomyocytes biorxiv.org/content/10.110…

Congrats Ajay Nadig on TRADE out now in Nature Genetics: nature.com/articles/s4158… These statistical metrics enable more meaningful comparisons in Perturb-seq atlases. Also, now find the HepG2 and Jurkat Perturb-seq datasets on GEO GSE264667!


