Jesse Weller
@jssweller
PhD candidate, @USC_Physics and @qcb_usc, University of Southern California | ML in chemistry and biology | AI for drug discovery
ID: 1572281140165365761
20-09-2022 17:47:15
145 Tweet
510 Followers
768 Following




















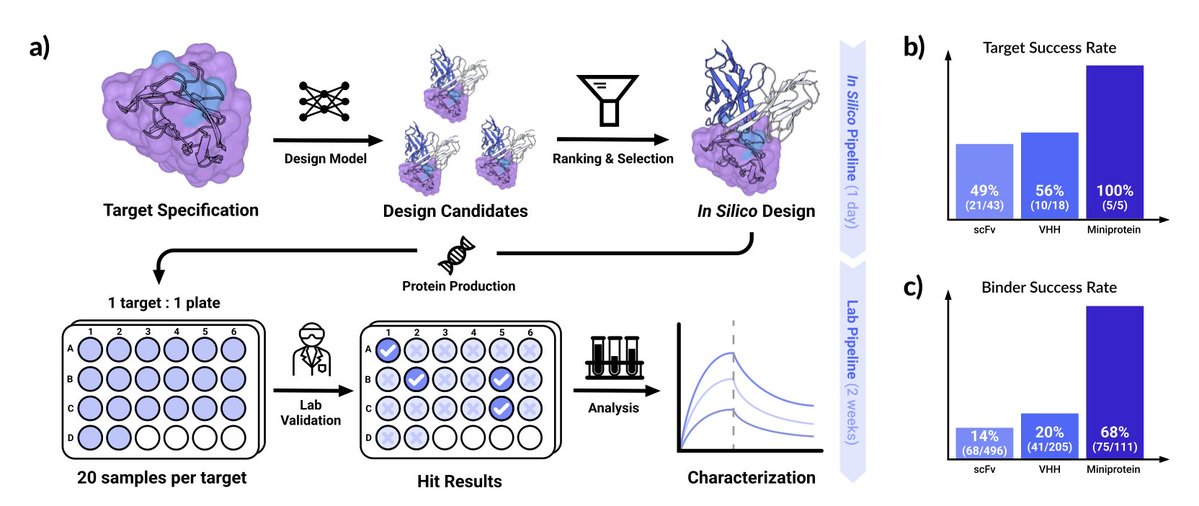
Zero-shot antibody design in a 24-well plate Chai Discovery 1. Researchers have introduced Chai-2, a multimodal generative model that marks a significant leap in de novo antibody and miniprotein design. This platform achieves an impressive 16% hit rate in de novo antibody