Kaido Lepik
@kaidolepik
Postdoctoral researcher in statistical genetics at the University of Lausanne. Developing causal methods.
ID: 2261878320
https://github.com/kaidolepik 25-12-2013 20:34:49
46 Tweet
75 Followers
93 Following


So happy to say this: our paper looking at the relationships between dietary patterns and blood metabolites is published! tinyurl.com/8cwwbn4c Thank you so much to everybody involved (Nicola Pirastu, Krista Fischer, Hanna-Kristel Valge, Tõnu Esko, Andres Metspalu, Jim Wilson)!

Very happy to see BayesRR-RC, a statistical approach we developed for GWAS to improve SNP-heritability estimation, discovery, fine-mapping and genomic prediction, published in Nature Communications 🎉! disq.us/t/439cniy Dept of Computational Biology, Uni Lausanne Université de Lausanne ISTAustria

Fresh off the press: Latent Heritable Confounder-MR has found a home at nature.com/articles/s4146… 🙌🥳 Very proud of this work, couldn't have done it without Zoltán Kutalik and Ninon Mounier !

Hi everyone, Today, our paper on microbiome host genetics (🧬) is published in Nature Genetics! 🎉🎉🎉 A 🧵 on what we found with A.Zhernakova, Serena Sanna, estebanL, Alex Kurilshikov and Shixian nature.com/articles/s4158…


Happy to share this highly collaborative work led by @MaarjaLepamets (w Chiara Auwerx, Lude Franke, Kaido Lepik etc) on omics-informed CNV confidence assessment and its translation to improved association signals. biorxiv.org/content/10.110…

Happy to see how a seed planted at a "write a review" course by Christophe Dessimoz under the gifted hands of Chiara Auwerx & Marie Sadler turned into such a great publication!

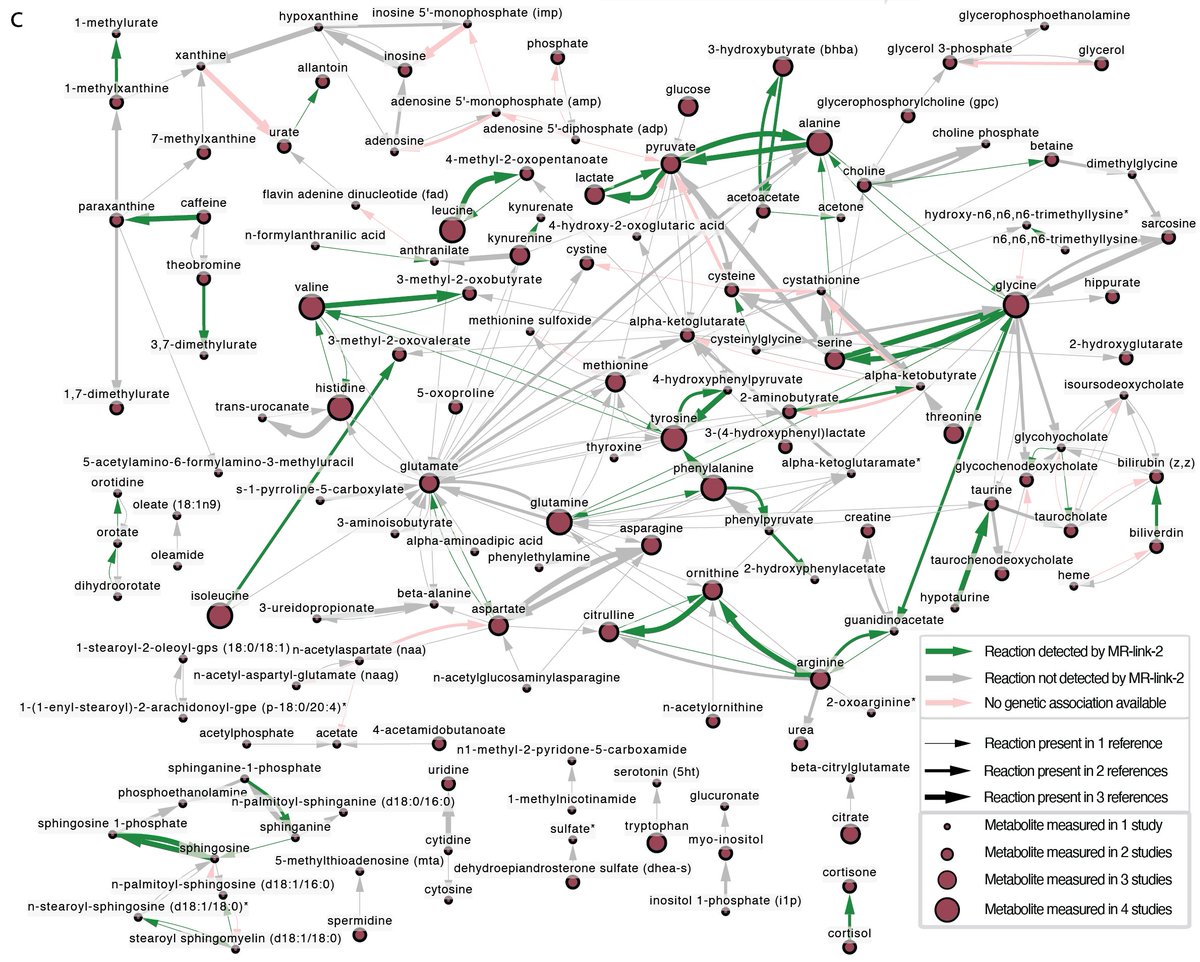
🔔New pre-print out!!! With Chiara Auwerx Marie Sadler Alexandre Reymond and Zoltán Kutalik we combined eQTL, mQTL and GWAS data in a multi-omics Mendelian Randomization approach to identify metabolites mediating transcript-trait associations. biorxiv.org/content/10.110…

Much of the knowledge on the 22q11.2 deletion syndrome stems from clinical cohorts. 🏥 Can we learn more by studying 22q11.2 CNVs in a population-based cohort such as the UK Biobank? 🌍 Find out in our new preprint by Malú Zamariolli! ⬇️ medrxiv.org/content/10.110…


So proud of Tabea Schoeler and Adriaan for their Dept of Computational Biology, Uni Lausanne public outreach video: youtu.be/NxnYZK3zczw

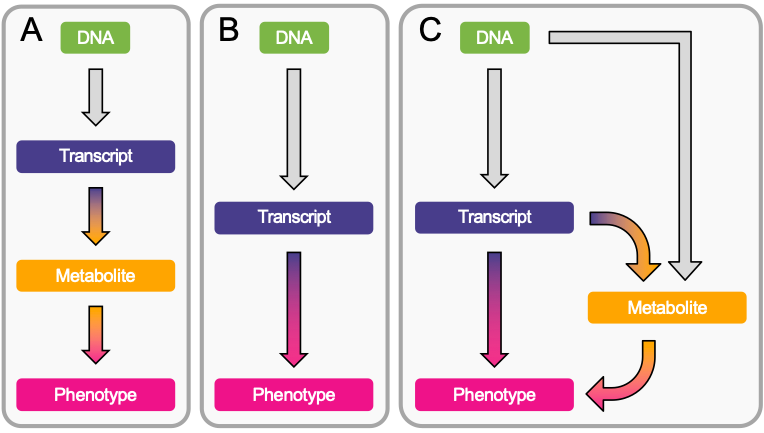
Excited to share that our paper about DNAm mediation through transcript levels is now published Nature Communications! nature.com/articles/s4146… Very grateful to all co-authors Chiara Auwerx Kaido Lepik Eleonora Porcu Zoltán Kutalik for their precious contribution! #omics #GWAS #MendelianRandomization


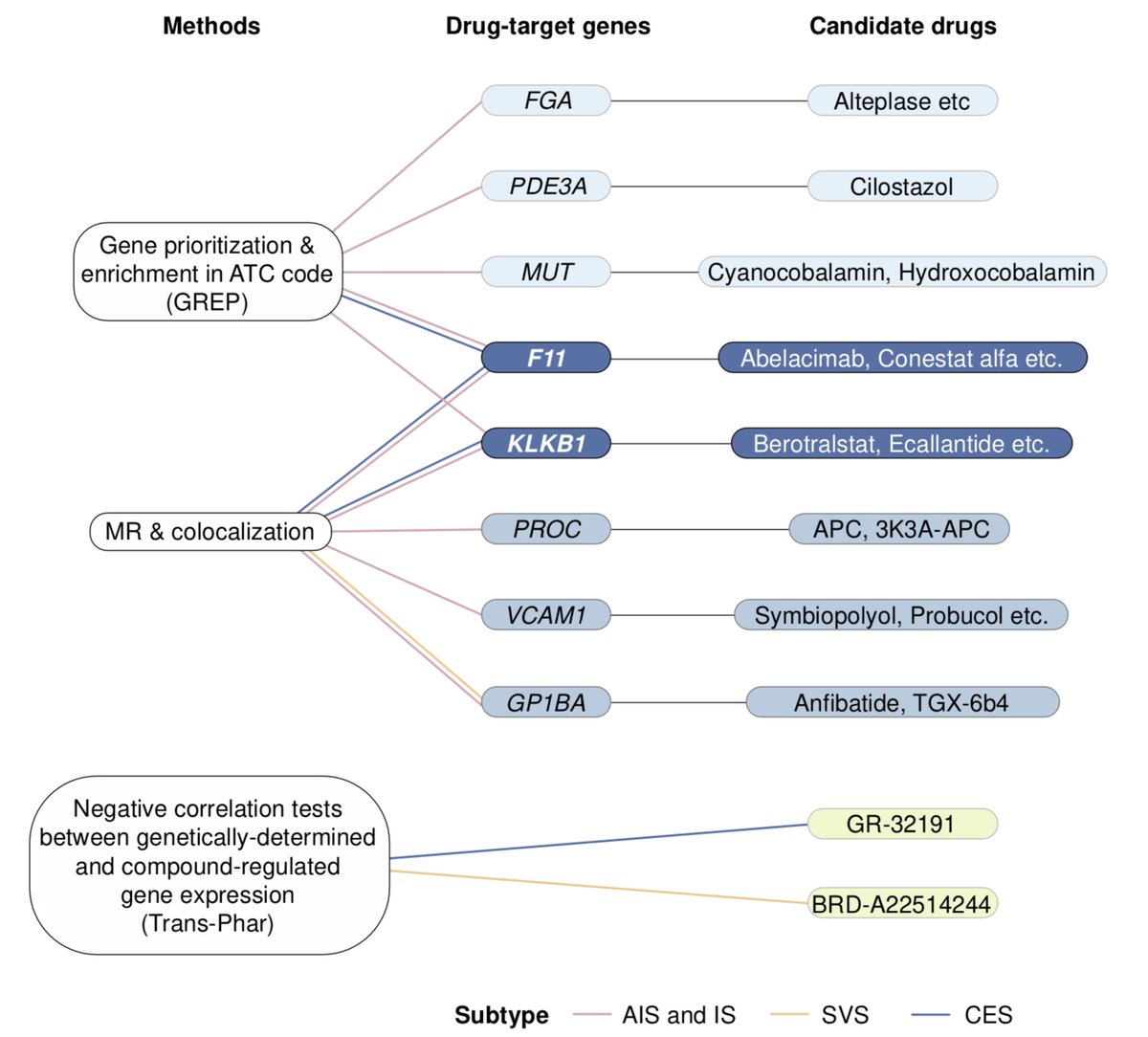
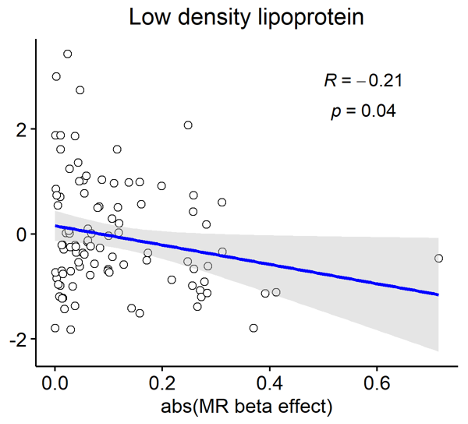
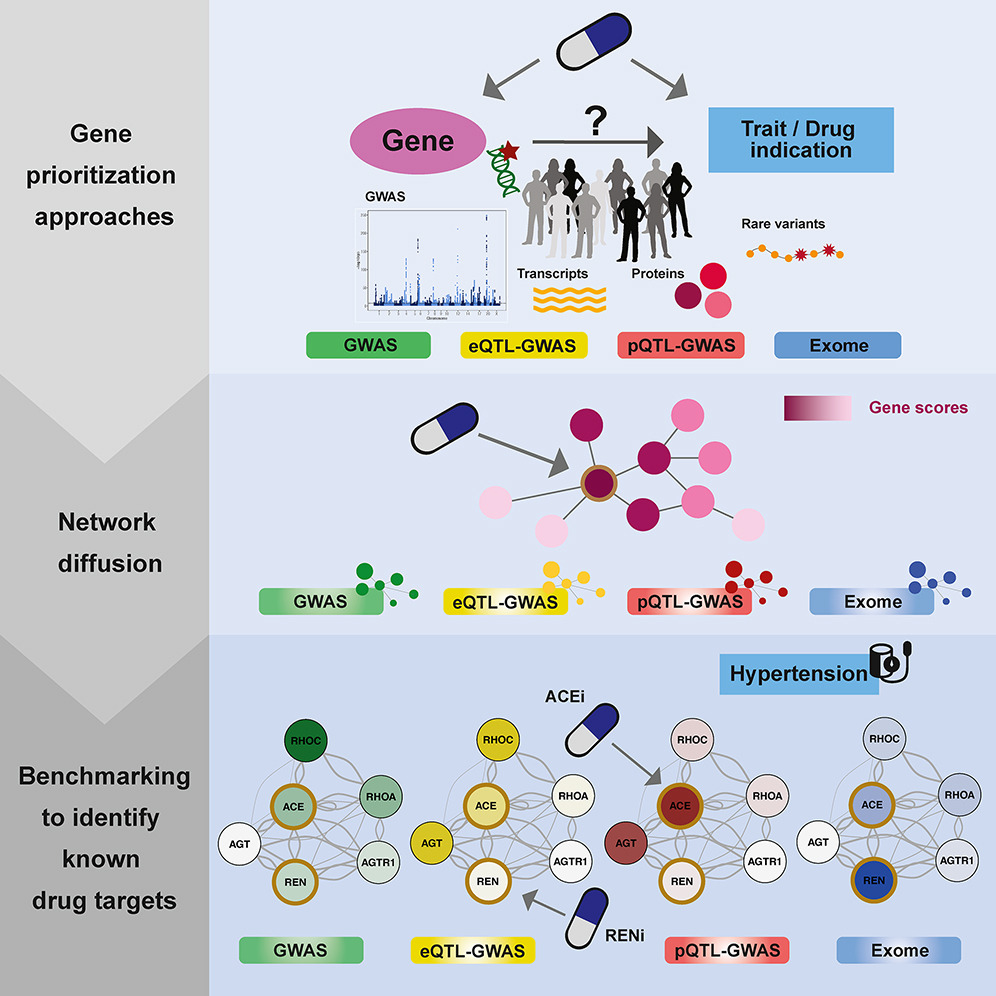
Most promising method to identify drug targets? 🎯 - #GWAS - #eQTL-GWAS - #pQTL-GWAS - #exome Excited to share our new preprint where we compare all methods including #network approaches (medrxiv.org/content/10.110…) Very grateful to all co-authors Zoltán Kutalik @cauwerx Patrick Deelen


Very happy to announce that our work “Multi-layered genetic approaches to identify approved drug targets” is now published Cell Genomics doi.org/10.1016/j.xgen… Many thanks to all co-authors Zoltán Kutalik @cauwerx Patrick Deelen for their precious contribution! 1/10

New year, new paper! 💫📄 Our study on the role of #CNVs as #pleiotropic modulators of #CommonDisease susceptibility is out in Genome Medicine: rdcu.be/dvonQ Major findings (👉🏻 x.com/CAuwerx/status…) + 3 of my favorite new insights based on reviewer’s comments in 🧵