Kateryna Tolmachova 🇺🇦
@kattlm
PhD candidate, Bode group, ETH Zürich
ID: 318516351
16-06-2011 16:21:38
152 Tweet
145 Followers
160 Following

One of my favorite quotes from today's talk by Frances Arnold. It resonated with me because I do it too often, only to be humbled by nature!

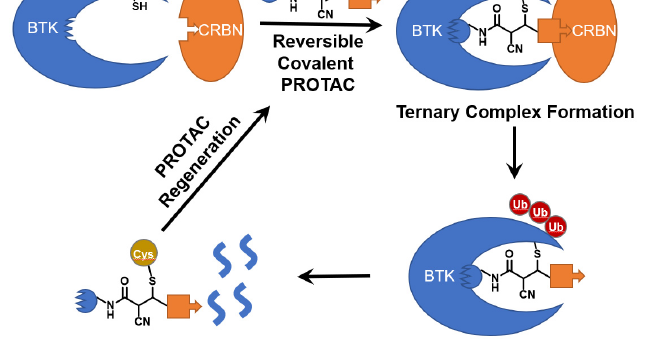
Our preprint on a new modality for targeted degradation of extracellular/membrane proteins is up on @ChemRXiv - a PROTAC for the extracellular space, so to speak. Looking forward to feedback. Inspired work led by postdoc Steven Banik, former PhD with Eric Jacobsen Harvard Chemistry.






So happy to see our UFM1 probes paper out in ACS Central Science! Jakob Farnung and I are big fans of hydrazides, which allow C-terminal protein modification easier than ever. Massive thank you to amosliang, Bode Group, Jacob Corn! pubs.acs.org/doi/10.1021/ac…

NEW #ASAP by Jeffrey Bode & team Bode Group UFM1 activity-based probes were developed via acylation of expressed C-terminal acyl hydrazides with carboxylic anhydrides. Probes showed remarkable selectivity toward UFMylation pathway enzymes go.acs.org/1eR


Bode Group adventures in autophagy: We report the presence of an unknown LIR motif in human ATG3 discovered using a combination of new activity-based probes, computational modeling and x-ray crystallography. The LIR motif is crucial for LC3-lipidation. biorxiv.org/content/10.110…
We love it when group members do projects we never expected to be working on...autophagy, protein-structures, CRISPR knockouts. Great work by Jakob Farnung and our collaborators Jacob Corn

This Edge article from Bode Group looks at the installation of electrophiles onto the C-terminus of recombinant ubiquitin and ubiquitin-like proteins! 👏 Sound interesting? You can read the paper in full, for free here🔗 ow.ly/whrS50M9A6C


Very happy to share work from my PhD in the Bode Group. We report the presence of an unknown LIR motif in human ATG3 discovered using a combination of new activity-based probes, modeling and x-ray crystallography. tinyurl.com/mrxtmsv6 ACS Central Science


Semisynthetic LC3 Probes for Autophagy Pathways Reveal a Noncanonical LC3 Interacting Region Motif Crucial for the Enzymatic Activity of Human ATG3 NEW #ASAP by Bode Group & co-workers Read it here: go.acs.org/56r


What does it take to degrade a protein? Turns out all you need is a C-terminal amide. In a great collaboration between Jacob Corn and Bode Group we identified C-terminal amides as a degron recognized by SCF/FBXO31. @matthias_muhar Raphael Hofmann

🎉Super excited to share our story on how the substrate receptor FBXO31 functions as a quality control factor by recognizing amides. This has been an amazing collaboration between Bode lab and Jacob Corn. Special shutout goes to @matthias_muhar nature.com/articles/s4158…



Chemoenzymatic Site-Specific Lysine Modification of Nanobodies and Subsequent Bioconjugation via Potassium Acyltrifluoroborate (KAT) Ligations | Journal of the American Chemical Society Jeffrey Bode Bode Group D-CHAB ETH Zurich ETH Zürich pubs.acs.org/doi/10.1021/ja…