Magnus Bauer
@kinasekid
Enjoying life one molecule at a time / Postdoc @UWproteindesign / ex @Stanford / PhD @LMU_Muenchen / tweeting in English, thinking in Bavarian, coding in Python
ID: 401543837
https://mastodon.social/@kinasekid 30-10-2011 18:14:40
376 Tweet
438 Followers
1,1K Following

Excited to share our preprint “BoltzDesign1: Inverting All-Atom Structure Prediction Model for Generalized Biomolecular Binder Design” — a collaboration with Martin Pacesa, Zhidian Zhang , Bruno E. Correia, and Sergey Ovchinnikov. 🧬 Code will be released in a couple weeks




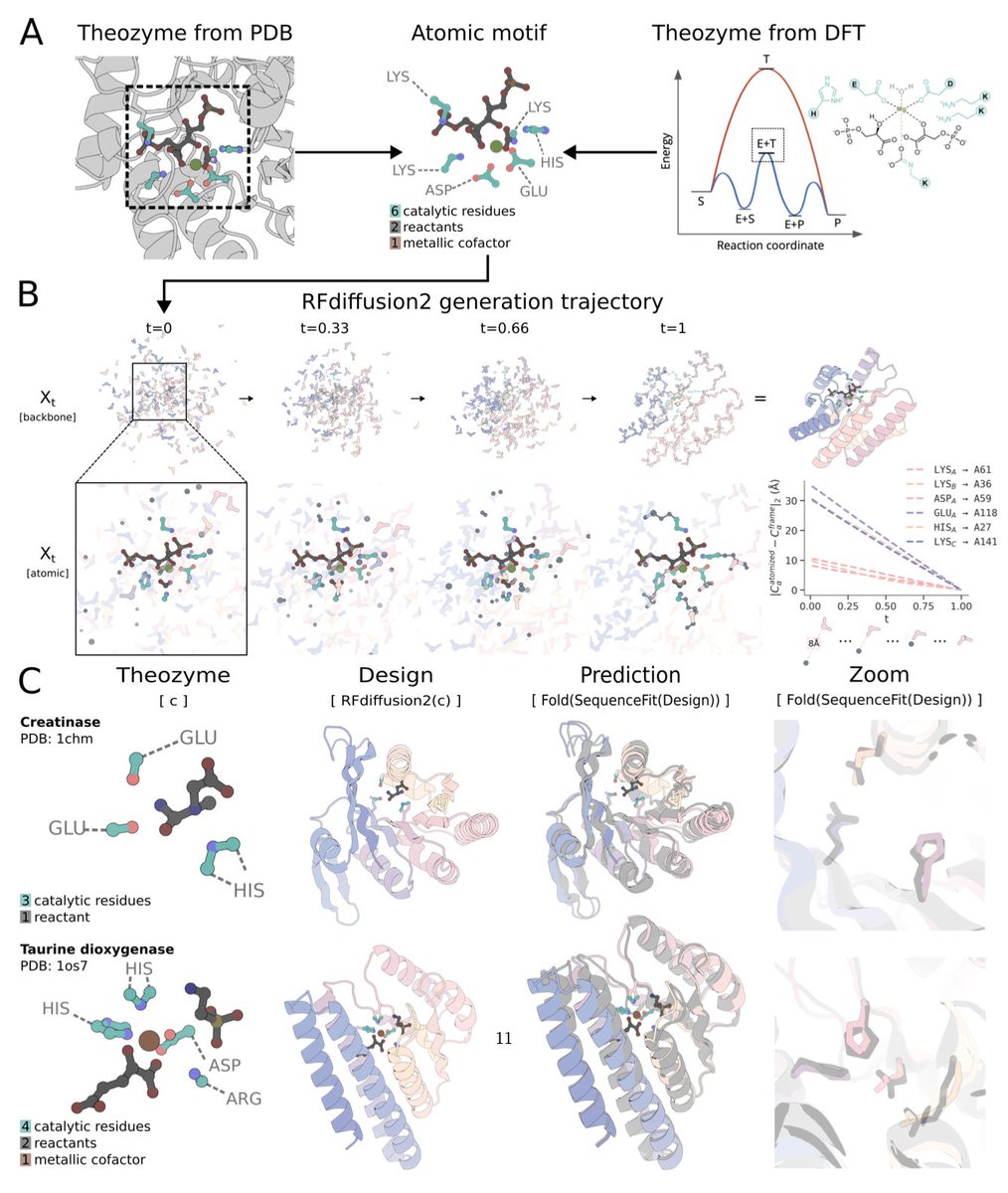
RFdiffusion2 generates new enzyme structures just from the most basic descriptions of their geometry. See Woody Ahern's thread to see the next generation of de novo enzyme design:



What could Alphafold 4 look like? (Sergey Ovchinnikov, Ep #3) 2 hours listening time (links below) To those in the (machine-learning for protein design) space, Dr. Sergey Ovchinnikov (Sergey Ovchinnikov) is a very, very well-recognized name. A recent MIT professor (circa early


Excited to share our latest releases to the FAIR Chemistry’s family of open datasets and models: OMol25 and UMA! AI at Meta FAIR Chemistry OMol25: huggingface.co/facebook/OMol25 UMA: huggingface.co/facebook/UMA Blog: ai.meta.com/blog/meta-fair… Demo: huggingface.co/spaces/faceboo…







🚀 Excited to release BoltzDesign1! ✨ Now with LogMD-based trajectory visualization. 🔗 Demo: rcsb.ai/ff9c2b1ee8 Feedback & collabs welcome! 🙌 🔗: GitHub: github.com/yehlincho/Bolt… 🔗: Colab: colab.research.google.com/github/yehlinc… Sergey Ovchinnikov Martin Pacesa