
Sayeh Gorjifard, MS, PhD
@lampoona
A down-to-mars girl. PhD Genome Sciences @uwgenome | MS Drug Development @JohnsHopkins | Chemistry + Art @dartmouth
ID: 1046749530
http://www.sayehgorjifard.com 30-12-2012 04:02:16
917 Tweet
238 Followers
1,1K Following

Thrilled to present our collaboration w/ Stergachis Lab members “The regulatory potential of transposable elements in maize”. Single-molecule Fiber-seq - accurate and sensitive accessibility in maize. New open chromatin regions in LTRs and much more! biorxiv.org/cgi/content/sh…



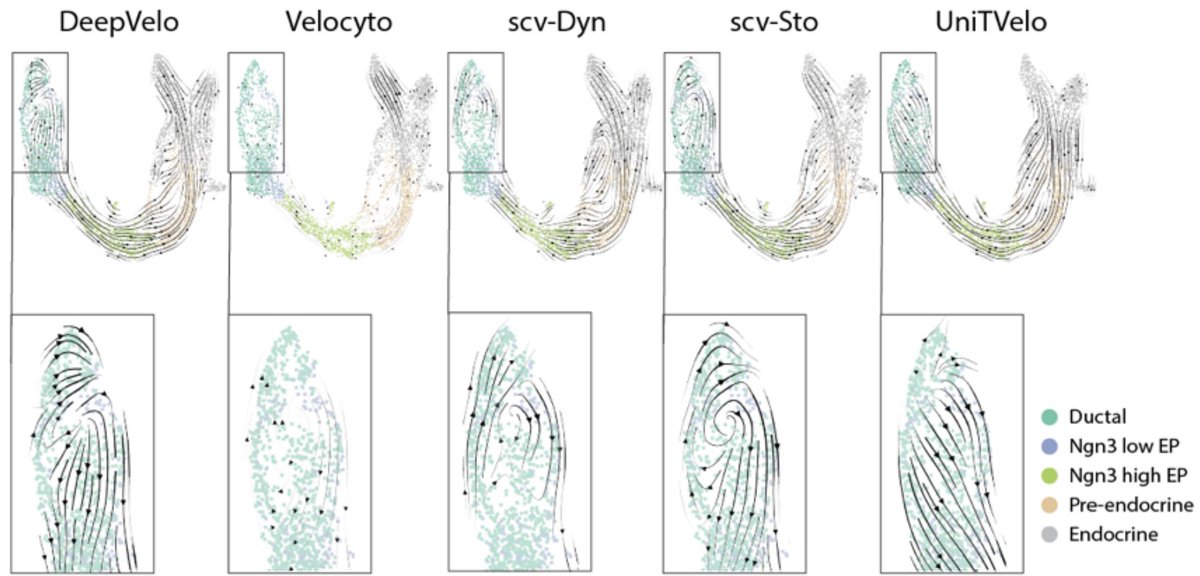
This recently published figure by Sarah Ancheta et al. is very disturbing and should lead to some deep introspection in the single-cell genomics community (I doubt it will). It demonstrates complete disagreement among 5 widely used "RNA velocity" methods 1/

Thrilled to share that our latest work in the Christodoulou lab is now published in Nature! nature.com/articles/s4158… Thanks to everyone who contributed Ivana Bukvin, Sammy Chan, Tomek Wlodarski, Angelo M Figueiredo , Chris Waudby 🦋, Lisa Cabrita, Anais Cassaignau, John Christodoulou!



Happy to share our paper 'Predicting Conformational Ensembles of Intrinsically Disordered Proteins: From Molecular Dynamics to Machine Learning' just published in @jphyschem Letters. Excellent work by J Aupic, Pavlína Pokorná Sharon Ruthstein read here: pubs.acs.org/doi/10.1021/ac…


Nick Desnoyer One movie of anther opening in A. thaliana 😉 Captured by spinning-disk microscope.


Dear Google Maps, please add a feature that maximizes a walking path for tree density, aka shade, and not time or distance. This would be great for anyone in a new city!













