Lorin Crawford
@lorin_crawford
Principal Researcher, @MSFTResearch
ID: 874910671133081601
http://www.lorincrawford.com 14-06-2017 08:45:33
977 Tweet
2,2K Followers
601 Following


This project was completed during an amazing internship at Microsoft Research New England with the invaluable guidance of Lorin Crawford and Ava Amini as part of the Ex Vivo project, with collaborators at Broad Institute Peter Winter and Sri Raghavan. Amazing science, amazing mentorship, with a view!



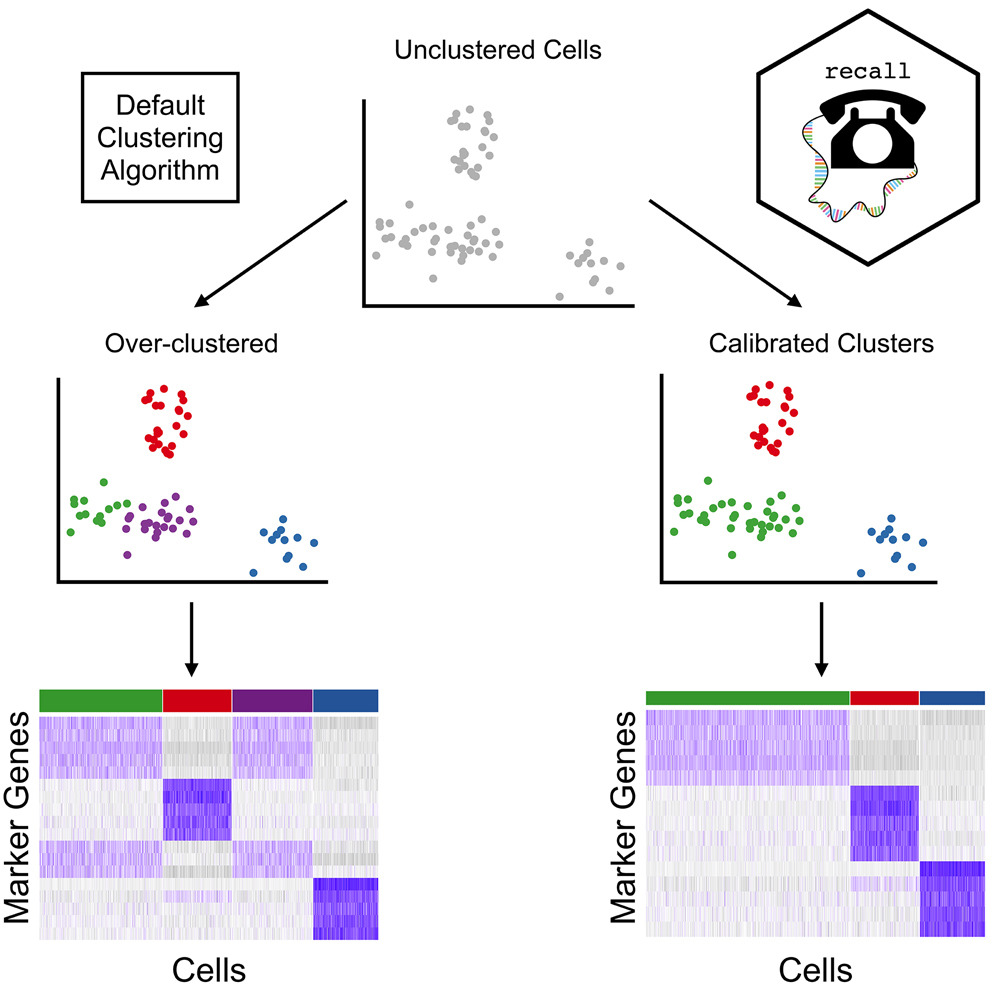
Great to see our paper presenting recall, a framework which calibrates clustering for the impact of data "double-dipping" in single-cell studies, out today in AJHG! Congratulations, Alan DenAdel and co-authors!

How can artificial variables improve clustering in single-cell RNA sequencing?Cell Press "Artificial variables help to avoid over-clustering in single-cell RNA sequencing" - Recent advances in single-cell RNA sequencing (scRNA-seq) have enabled large-scale transcriptomic
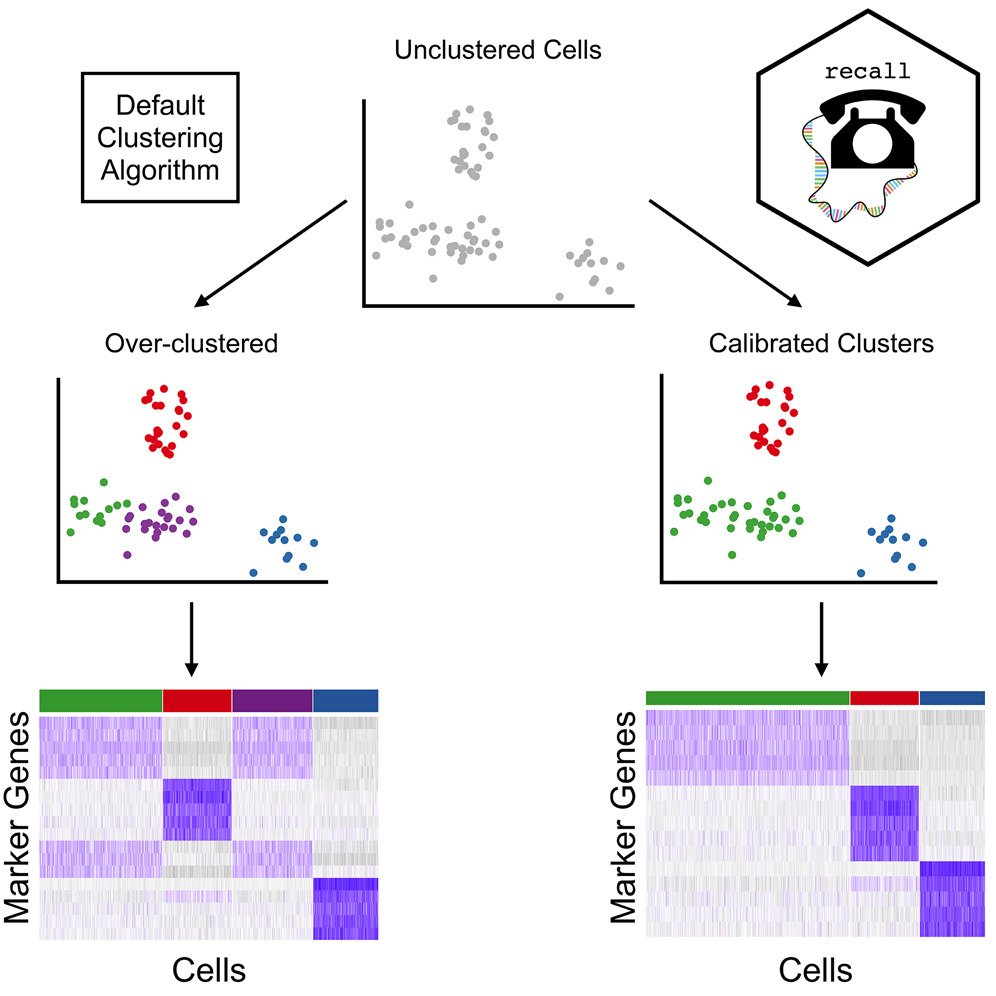

Over-clustering of single-cell RNA-seq data can produce spurious results. Alan DenAdel, Lorin Crawford, & co of AJHG latest study introduce recall, a new method, protecting against over-clustering & enables rapid analysis of single-cell RNA-seq data: cell.com/ajhg/abstract/…




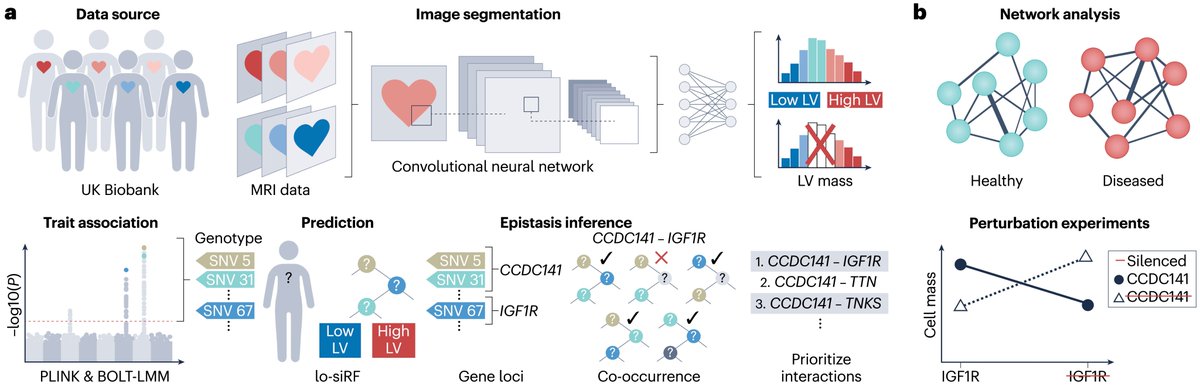
Our paper has been published Nature Genetics! Through new statistical methods, we shed light on fundamental questions about cellular response to genetic perturbations. Our work is a substantial advance towards rigorous characterization and comparison of massive perturbation atlases.

🎉 Congratulations to Eric and Wendy Schmidt Center postdoctoral fellow Sebastiano Cultrera di Montesano and his team on their latest preprint! Learn more: doi.org/10.1101/2025.0…

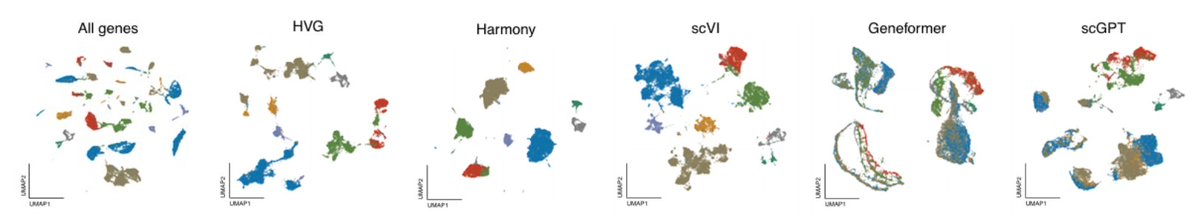
How well do single-cell foundation models perform w/o finetuning? Our work Genome Biology shows that in zero-shot settings, scGPT and Geneformer often underperform traditional methods, raising questions about their utility for biological discovery. 📰 genomebiology.biomedcentral.com/articles/10.11…




So awesome to see this finally out—huge congrats to all the authors! If interested, we covered the highlights in a recent News & Views piece in Nature Cardiovascular Research (s/o to Julian Stamp for jumping in as co-author) 🔗 rdcu.be/epxFW