
Andreas Loukas
@loukasa_tweet
machine learning biology. doesn't work on LLMs yet. likes to think about how machines learn
ID: 636167919
15-07-2012 10:32:51
243 Tweet
1,1K Followers
487 Following

Really excited to be introducing our new framework for de novo protein design, tomorrow at the MLDD_Workshop #ICLR_2022 at 10:30-10:50am ET! Paper: openreview.net/forum?id=DwN81…

Highly recommended post-doc and PhD positions on generative models for discrete data with Alexandros Kalousis 👇

In this Tuesday's reading group session, we discuss 'SPECTRE : Spectral Conditioning Helps to Overcome the Expressivity Limits of One-shot Graph Generators' with the authors Karolis Martinkus and Andreas Loukas!🤗 11am EST / 5pm CEST in the Zoom here: hannes-stark.com/logag-reading-… 1/2



Very happy for this work and collaboration with Nikos Karalias, Josh Robinson, and Stefanie Jegelka

I am hiring a #machinelearning scientist on my team @Prescient Genentech please apply if interested in geometric deep learning, generative modeling, de novo protein engineering, and want to be on the interface of structure/ML/experiments/engineering roche.wd3.myworkdayjobs.com/ROG-A2O-GENE/j…
Next Tuesday Oct 10th @ 4 pm EST we'll have Karolis Martinkus present AbDiffuser: Full-Atom Generation of In-Vitro Functioning Antibodies Zoom links will be emailed on the day before! Read the paper here: arxiv.org/abs/2308.05027 Karolis Martinkus Prescient Design

Karolis and Andreas Loukas's excellent talk at Cambridge Computer Science on de novo antibody design with diffusion models is now on youtube. Thank you for the excellent discussions! youtube.com/watch?v=9hU2rn… cc. Prescient Design

LoG 2023 is less than one month away, and we're super excited to hear from our keynote speakers, Andreas Loukas Jure Leskovec Kyle Cranmer and Stefanie Jagelka! logconference.github.io


Will be at NeurIPS Conference as well! Reach out if you want to talk


This was quite a fun experience for me. A unique forum to express thoughts that don't fit within usual publication mediums. Thanks Michael Galkin and Michael Bronstein for organizing us



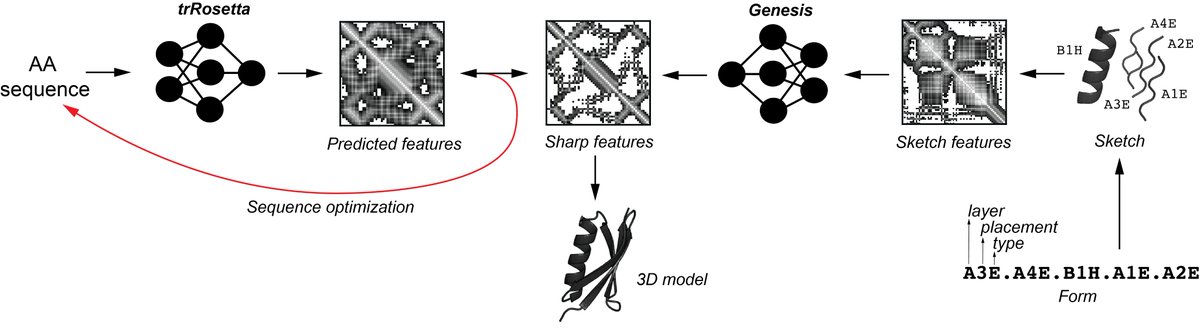
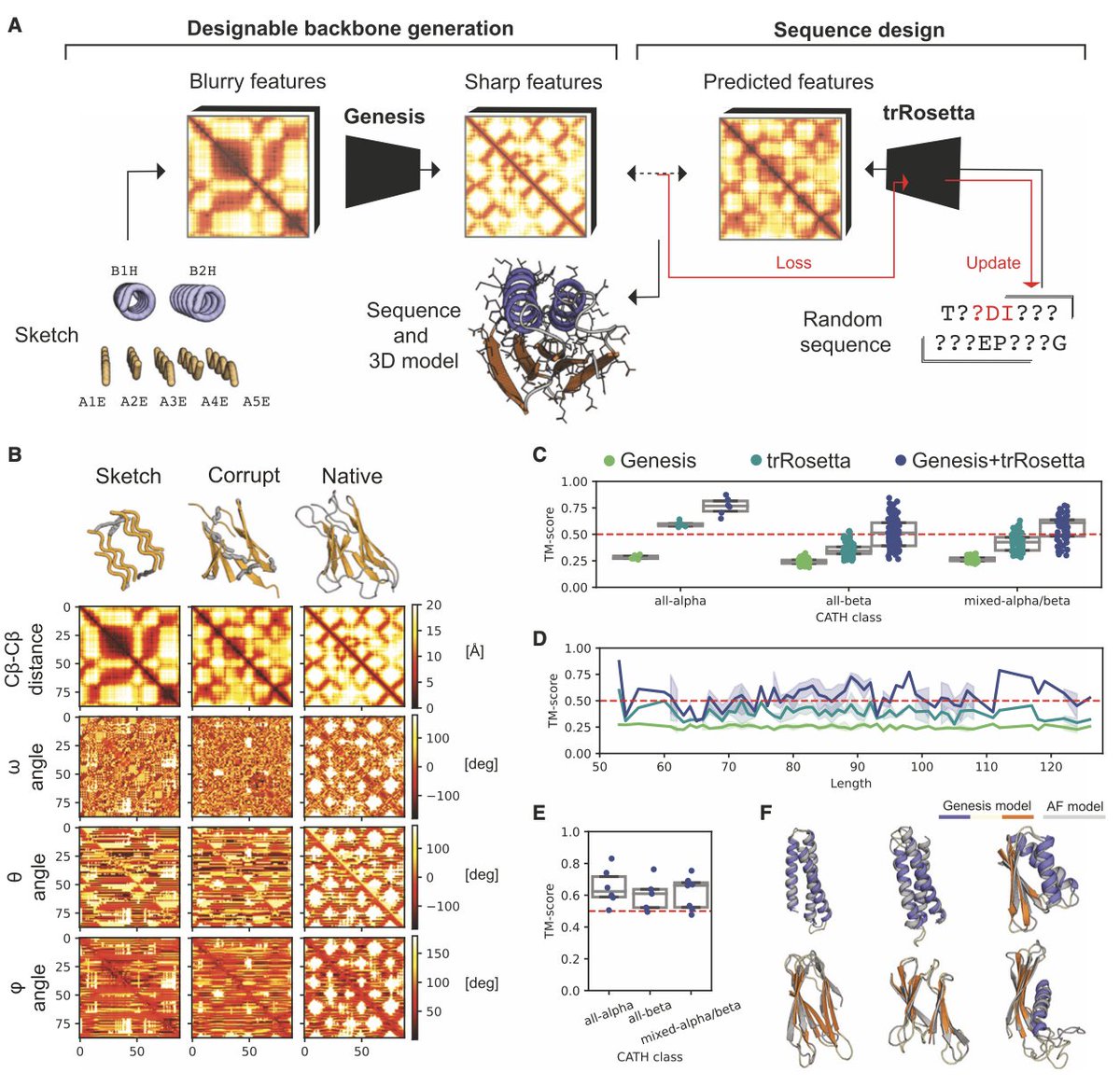
Exploring “dark-matter” protein folds using deep learning Cell Systems • Introducing Genesis VAE, a convolutional variational autoencoder that transforms low-resolution protein fold sketches into designable, stable 3D models. • Genesis VAE enables rapid exploration of

The last 3 years have been nothing short of amazing. I am grateful to have been immersed in drug design amongst wonderful colleagues 💊 In 2025, I will be starting a new chapter at Isomorphic Labs. I am excited to join this talented team and to embark on this new adventure 🚀


