Marton Olbei
@martonolbei
Cytokine Networks | Research Associate @KorcsmarosLab @imperialcollege | cytokinelink.org | 🦋 : martonolbei.bsky.social
ID: 875277400099180545
15-06-2017 09:02:48
1,1K Tweet
309 Followers
717 Following

For the 11th times, we delivered a #NetworkMedicine & Cytoscape training EMBL-EBI Training's Multiomics course. Great questions, smart participants & research-driven teaching atmosphere EMBL-EBI We covered OmniPath + use cases we've done Metabolism, Digestion & Reproduction & Earlham Institute W/ Marton Olbei




Great news! 🍾 We are excited to host + support the HFSP fellowship of Inez Roegiers at Metabolism, Digestion & Reproduction Imperial Medicine This project to bioprint microbes & #organoids will have revolutionary impact on how we study host-microbe interactions & on the role of the #microbiome in #IBD! 👏

Final program for this year's Signaling Workshop is live! 2025.signalingworkshop.org/program.php 🎟️ Only 10 spots left! 💡Unique mix of speakers rarely at the same event 🌍Topics: #IBD, #Microbiome, #Organoids,#SystemsBiology, #PrecisionMedicine 🔗Register till 28 April: signalingworkshop.org




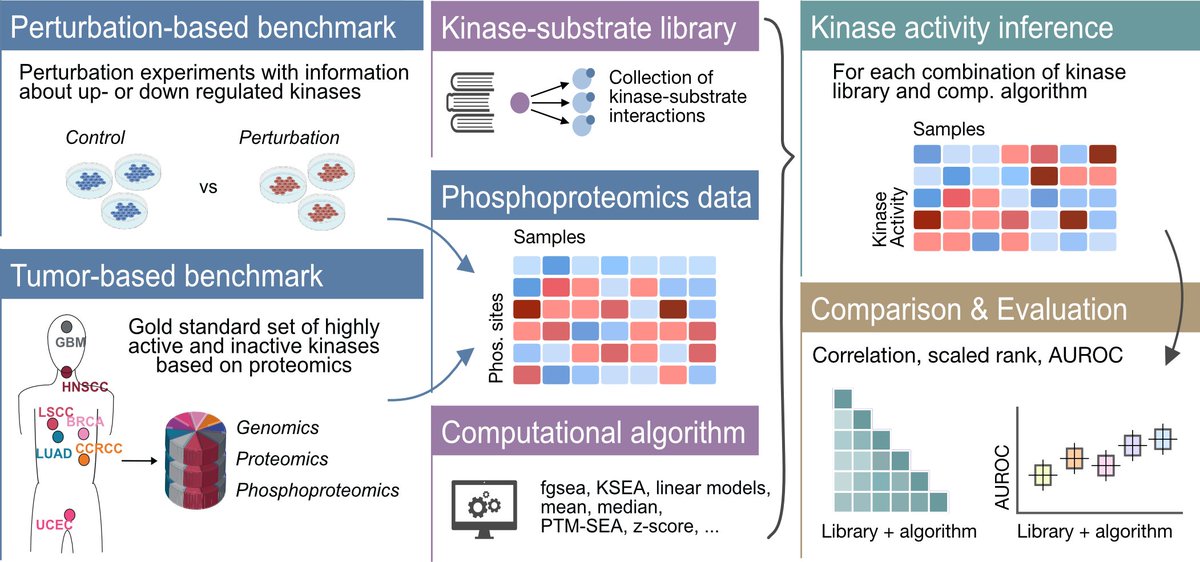
Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭Our revised comprehensive evaluation of kinase activity inference tools, done in collaboration with the Zhang lab BCMHouston, is now out Nature Communications 🔬 tinyurl.com/4twuc6z4

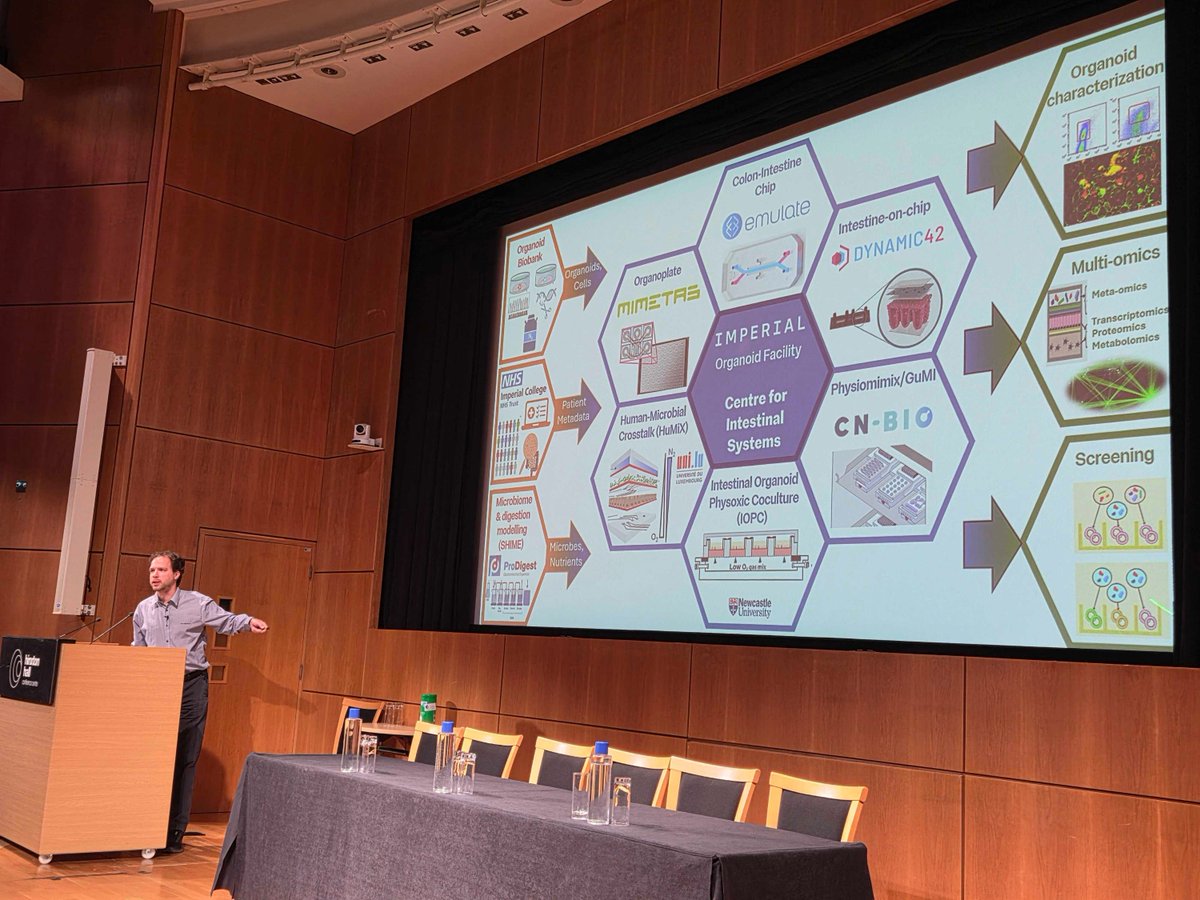
Inspiring few days on how to identify mechanisms for #MyalgicEncephalomyelitis (ME) / Chronic Fatigue Syndrome. #SystemsBiology session w/ modoslab #Cytokines w/ Marton Olbei #organoids & new #Gutonchip + #microbiome platforms ImperialOrganoid Metabolism, Digestion & Reproduction Thanks for @invest_in_me (Invest in ME Research) 🇪🇺


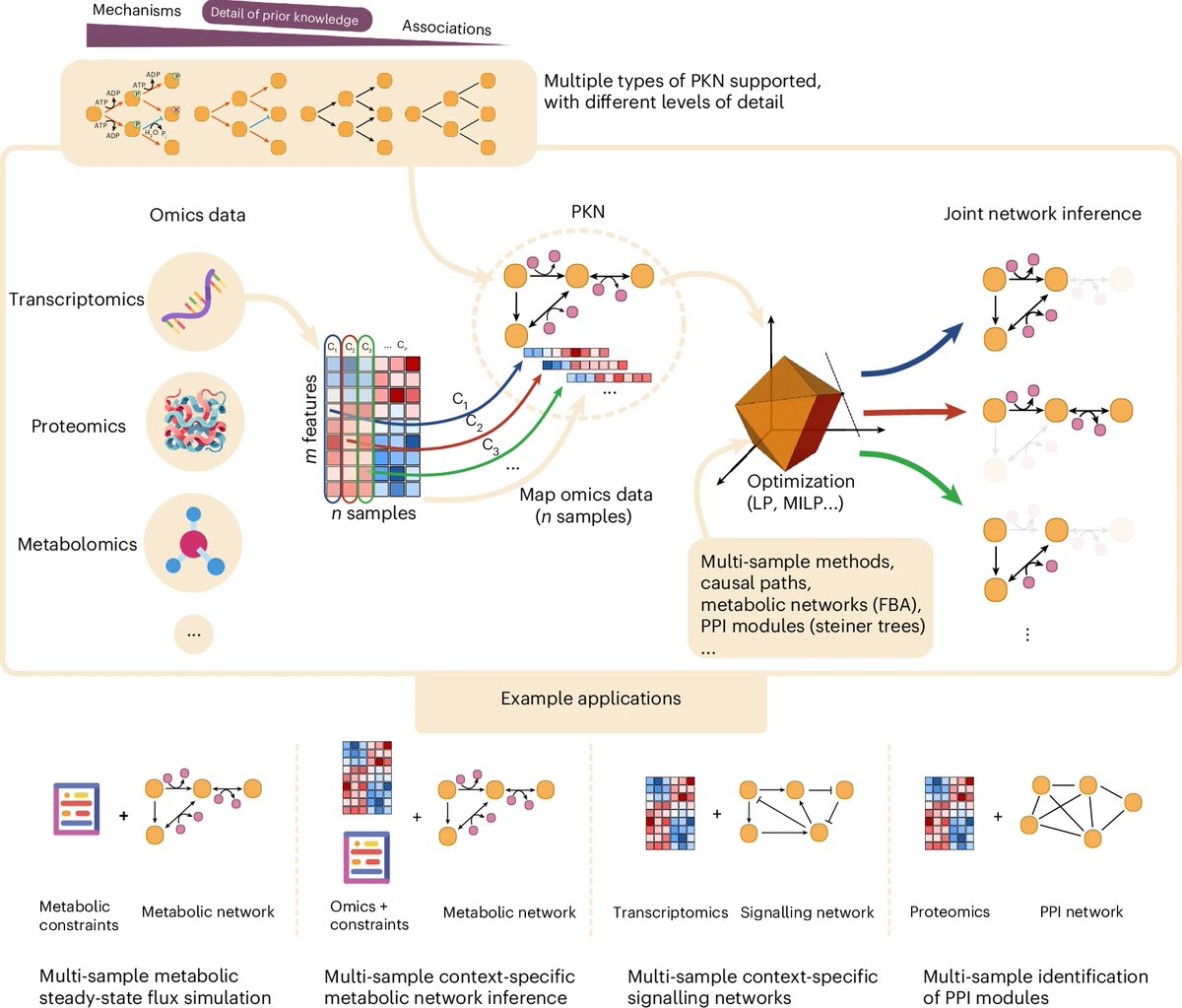
In Nature Biotechnology, CDS-affiliated Itai Yanai and Martin Lercher discuss how openness fuels discovery. They highlight the evolutionary nature of research, where ideas evolve through variation and selection, and advocate for embracing a creative process. nature.com/articles/s4158…

We’ve been named as 1st in the UK and 2nd in the world by QS QS World University Rankings! This recognises Imperial as one of the best places to study, research, work and grow. Thanks to everyone in our community for making this possible 💙 #QSWUR




Great to see this work from Shiv Radhakrishnan and colleagues out in #GutMicrobes - using network analysis to explore #microbiome #metabolome interactions in an #IBD inception cohort: doi.org/10.1080/194909… Imperial Hepatology Metabolism, Digestion & Reproduction KorcsmarosLab

This #GutMicrobes paper has been a nice collaboration w/ Shiv Radhakrishnan, James Alexander & Prof Horace Williams. Our side was led by Marton Olbei. Great to finally analyse omics data of a real inception #IBD cohort to capture meaningful signals/markers not masked by therapy.