Matteo Di Bernardo
@mat10_d
PhD student @MITCSBPhD with @iaincheeseman, previously @FulbrightPrgrm Nigeria, @columbiacancer
ID: 2181049021
07-11-2013 23:25:57
187 Tweet
435 Followers
1,1K Following

In collaboration with Reuben Saunders, Jonathan Weissman's Lab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…

They're coming fast and furious! Also today Science Magazine add EVOLVEpro, a protein large language model combined with a regression model, for genome editing, antibody binding and more applications for evolving proteins science.org/doi/full/10.11… Omar Abudayyeh Jonathan Gootenberg Kaiyi Jiang

How can foundational models + directed evolution make proteins >100x-fold better? We (Omar Abudayyeh, Kaiyi Jiang, Matteo Di Bernardo, Zhaoqing Yan) report in Science Magazine on EVOLVEpro, a general active learning approach to improve protein and enzyme function science.org/doi/10.1126/sc…



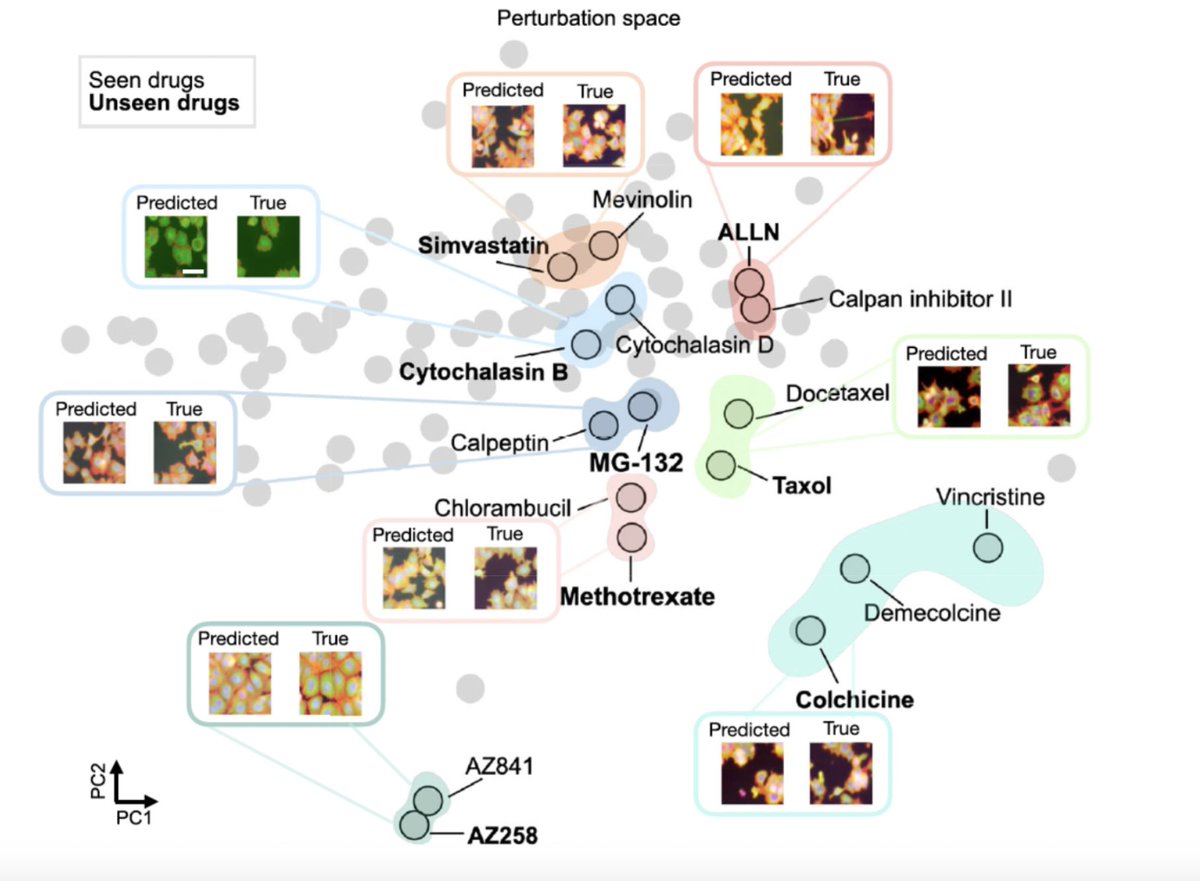
(1/8) IMPA is published now in Nature Communications. (a) It can generate phenotypic cell painting/microscopy data images under unseen drug and genetic perturbations. (b) It learns a perturbation map of treatment similarities (small molecules together with genetic) and (c) it corrects




Check out Whitehead Institute's symposium – AI: Advancing Foundational Biology – today (Tuesday, April 8), from 2-5:00 p.m., followed by a poster session and reception at 5 p.m. Eric and Wendy Schmidt Center Director Caroline Uhler will be keynoting at 2:15 p.m. Learn more: calendar.mit.edu/event/ai-advan…



🚫 No dyes. No bleaching. 🔬 Just AI + label-free microscopy = vivid virtually stained images New in Nature Machine Intelligence: A deep learning model that enables robust virtual staining across microscopes, cell types & conditions. #CZBiohubSF Shalin Mehta explains:

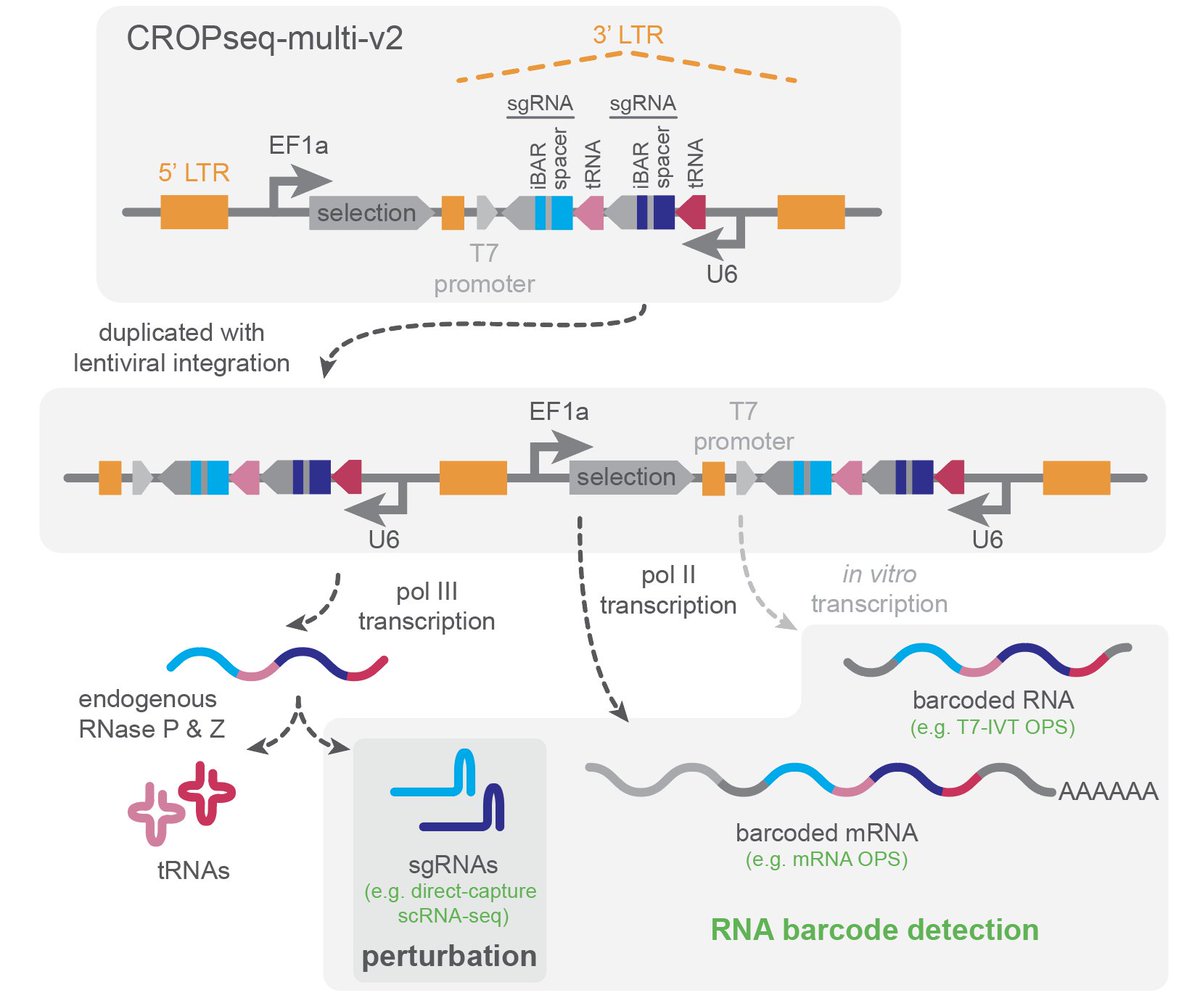
We’re Blainey Lab sharing a major update to CROPseq-multi, our versatile system for CRISPR screens that is compatible with individual and combinatorial perturbations, diverse SpCas9-based technologies, and multiple high-content, single-cell readouts. biorxiv.org/content/10.110…