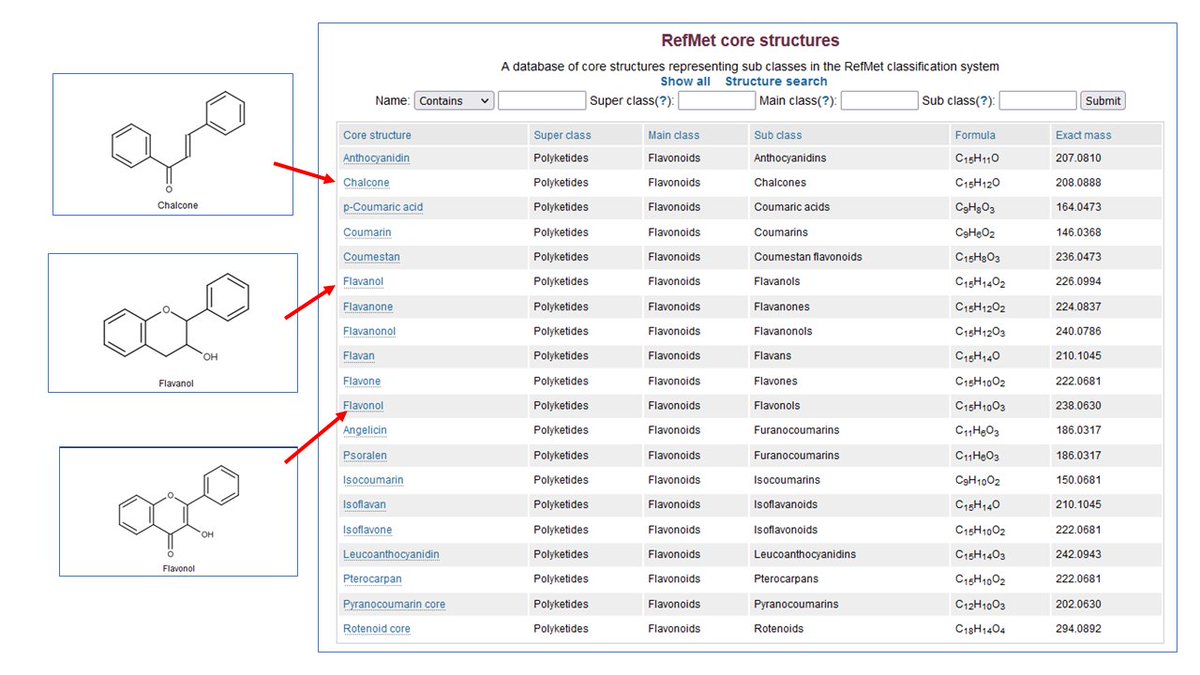
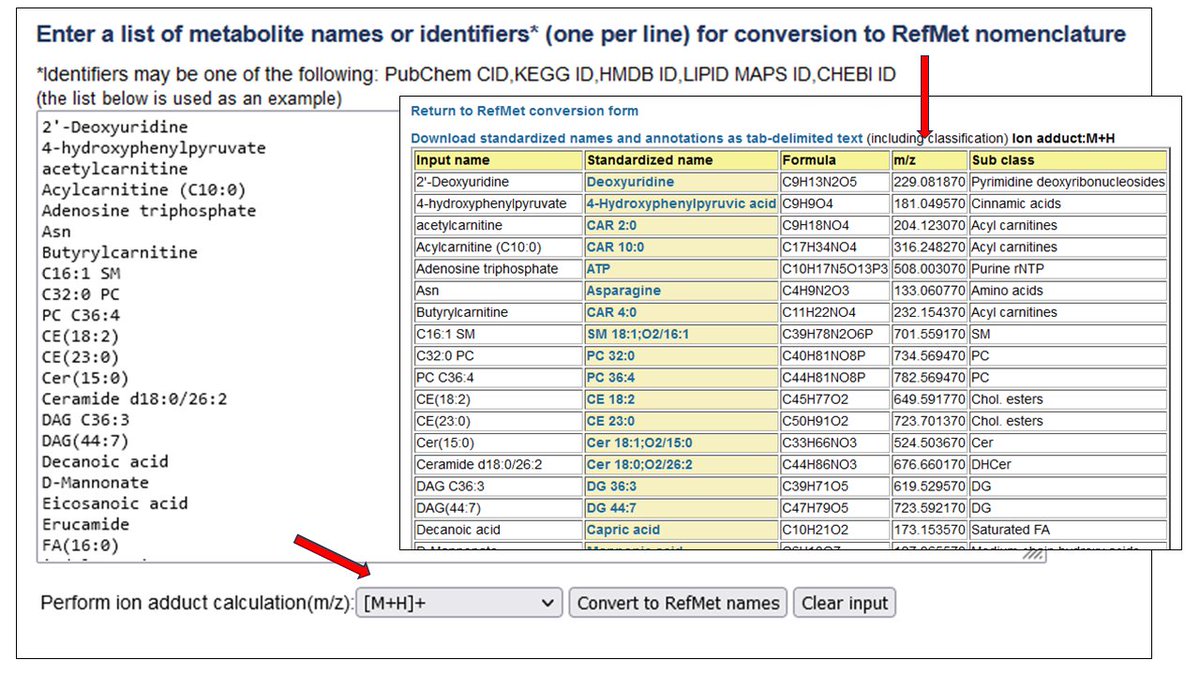
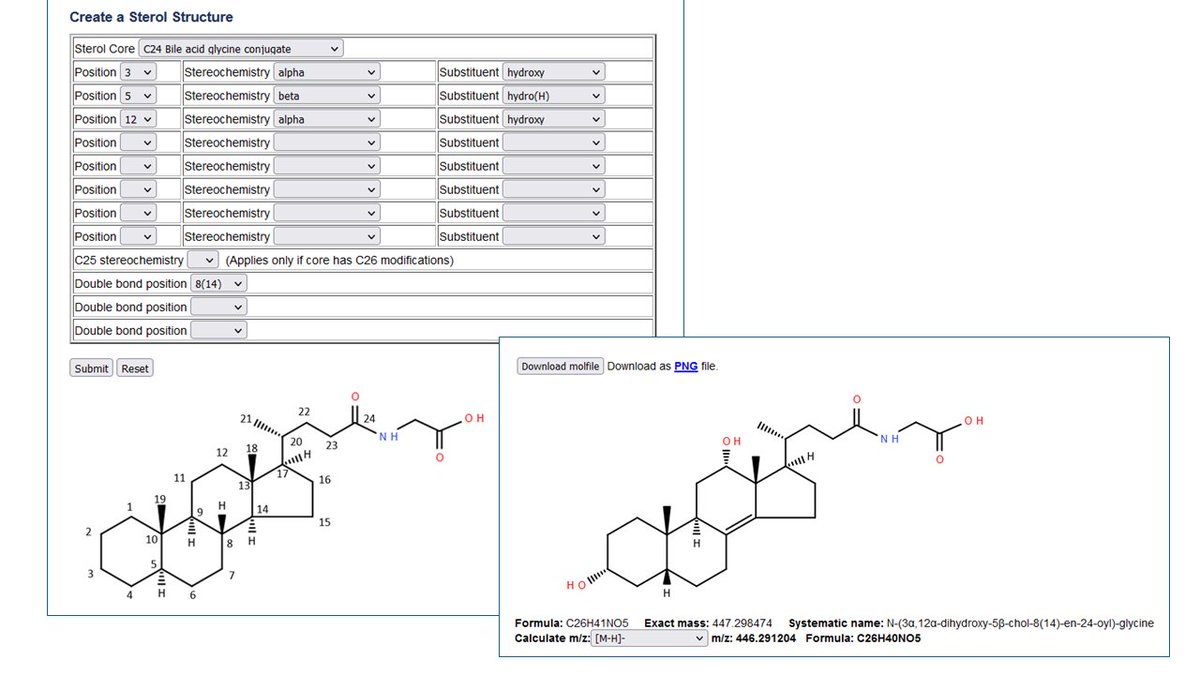
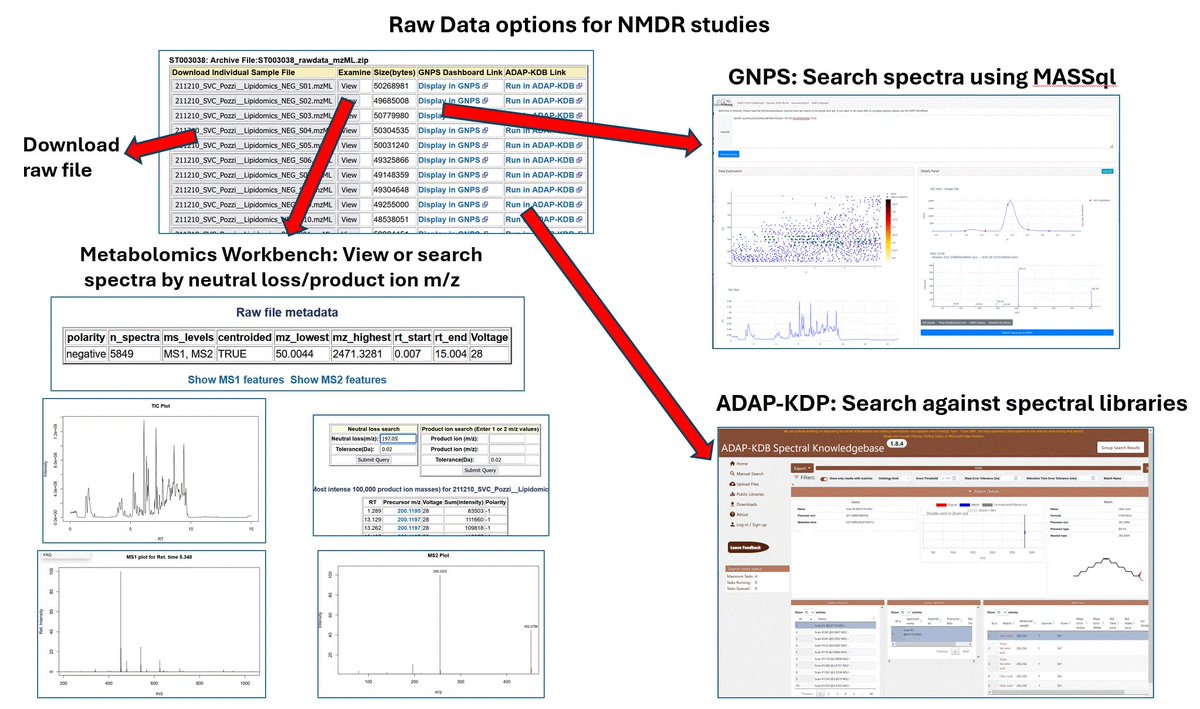
Metabolomics Workbench
@metabolomicswb
The Metabolomics Workbench at @UCSanDiego, funded by the @NIH_CommonFund | The National Metabolomics Data Repository (NMDR) and resource for metabolomics.
ID: 852600378616971264
http://www.metabolomicsworkbench.org 13-04-2017 19:12:25
603 Tweet
2,2K Followers
196 Following



The National Metabolomics Data Repository (NMDR) at Metabolomics Workbench has just processed its 2,000th study! MS/NMR data/metadata on studies covering over 130 species. Browse/search/download at metabolomicsworkbench.org


We're very happy to be the 2000th follower for Metabolomics Workbench as they reach the epic milestone of having curated their 2000th metabolomics dataset!

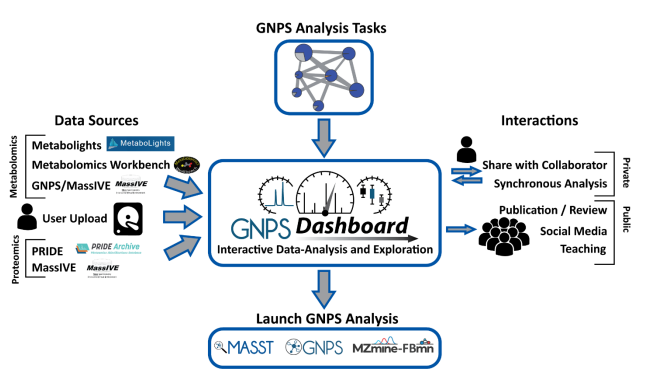
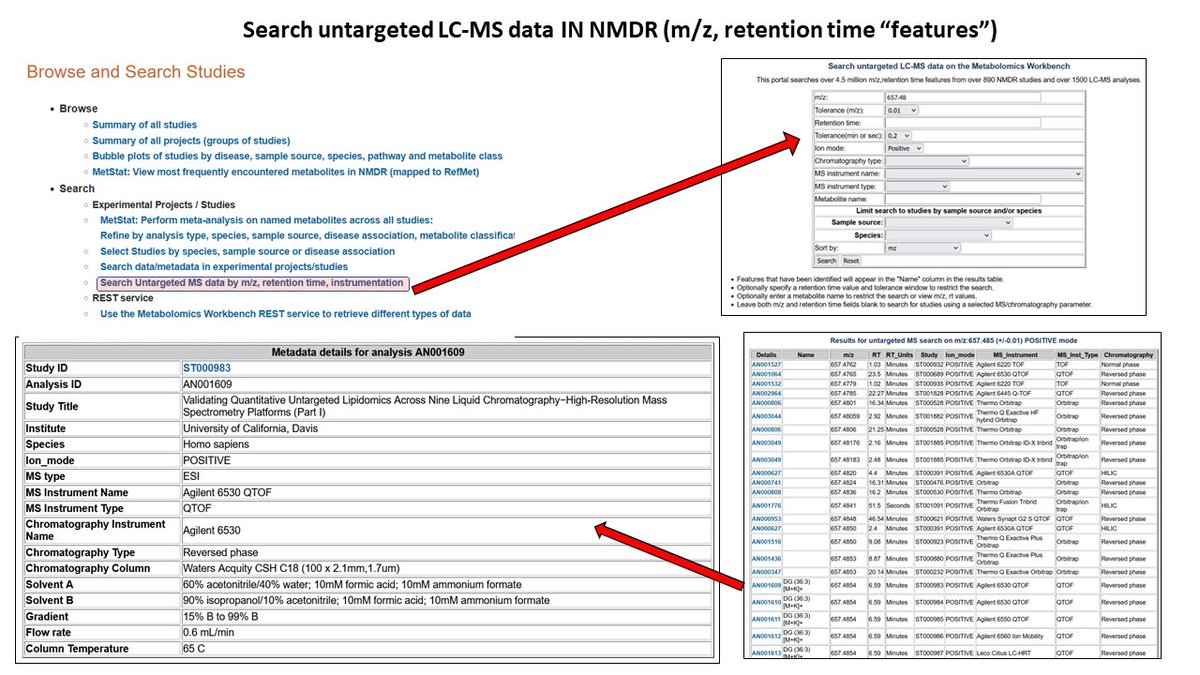
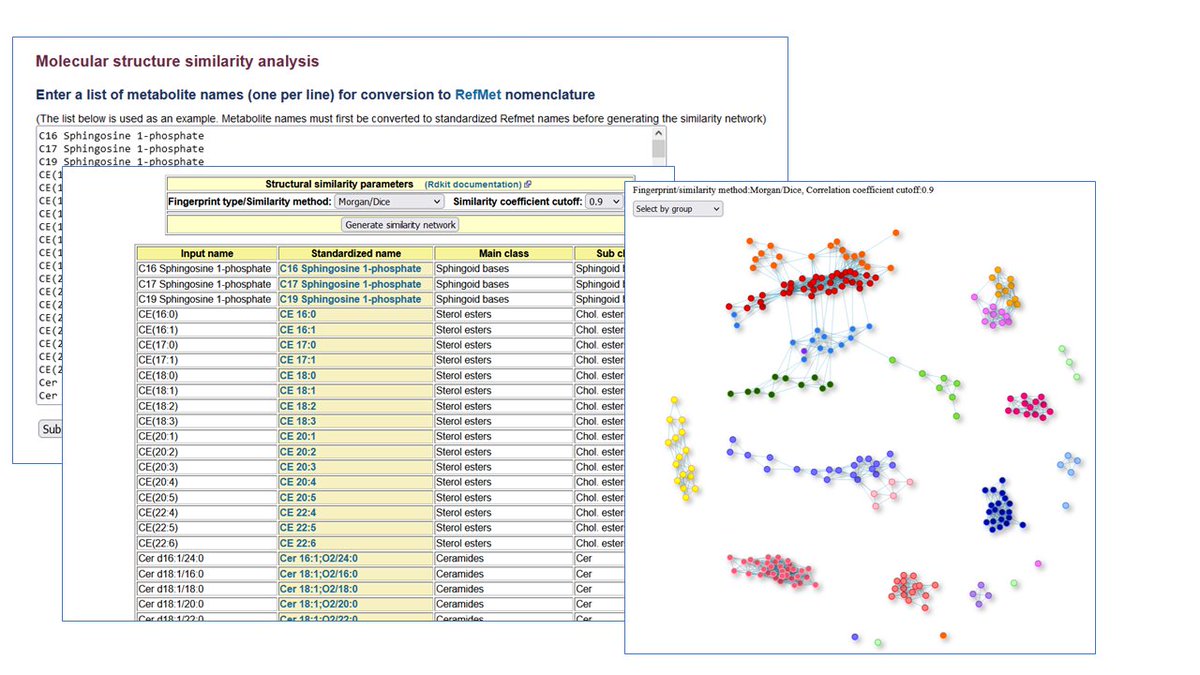
A new set of NMDR browsing/searching/analysis tutorials has just been added to Metabolomics Workbench at metabolomicsworkbench.org/data/tutorials…


The #METLIN ion mobility library is growing in a variety of ways, partly due to Cayman Chemical contributions, as well as other efforts. The plan is to make a 30+K version of #METLIN ion mobility downloadable.



Continue your #MANA2022 Day 3 with an Instructional Workshop discussing Metabolomics Workbench and the National Metabolomics Data Repository hosted by Eoin Fahy! Open science and data sharing are key to expanding our knowledge and capabilities in metabolomics!



Did someone mention "reference spectrum" Nature Methods? #METLIN is doing its best, now w 880K molecular standards (each w reference MS/MS data at 4 different collision energies). As always, thank you Cayman Chemical & @AvantiLipids doi.org/10.1038/s41592… doi.org/10.1038/s41592…






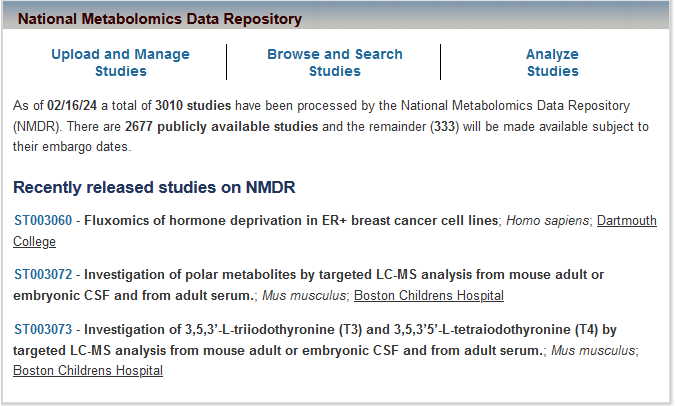
The National Metabolomics Data Repository (NMDR) at Metabolomics Workbench has just processed its 3,000th study! MS/NMR data/metadata/raw data on studies covering over 300 species. Browse/search/download at metabolomicsworkbench.org