Milena Pavlović
@milenapavl
Postdoc researcher at Sandve lab @UniOslo interested in machine learning, causal inference, simulation and their applications in biomedical sciences 📚💻📚
ID: 770376390428467201
https://www.mn.uio.no/ifi/english/people/aca/milenpa/index.html 29-08-2016 21:43:39
107 Tweet
637 Followers
860 Following






A 4-year PhD fellowship in Informatics/Bioinformatics is available at Centre for Bioinformatics, University of Oslo as part of @UiO_LifeSci. The projects will focus on synthetic data generation, applied machine learning and benchmarking within molecular life sciences: jobbnorge.no/en/available-j…. Please RT.









Our paper on the linguistics-based formalization of antibody language is out in Nature Computational Science. Work led by Mai Ha Rahmad Akbar and Philippe A. Robert . Summary thread: x.com/PRobertImmodel….

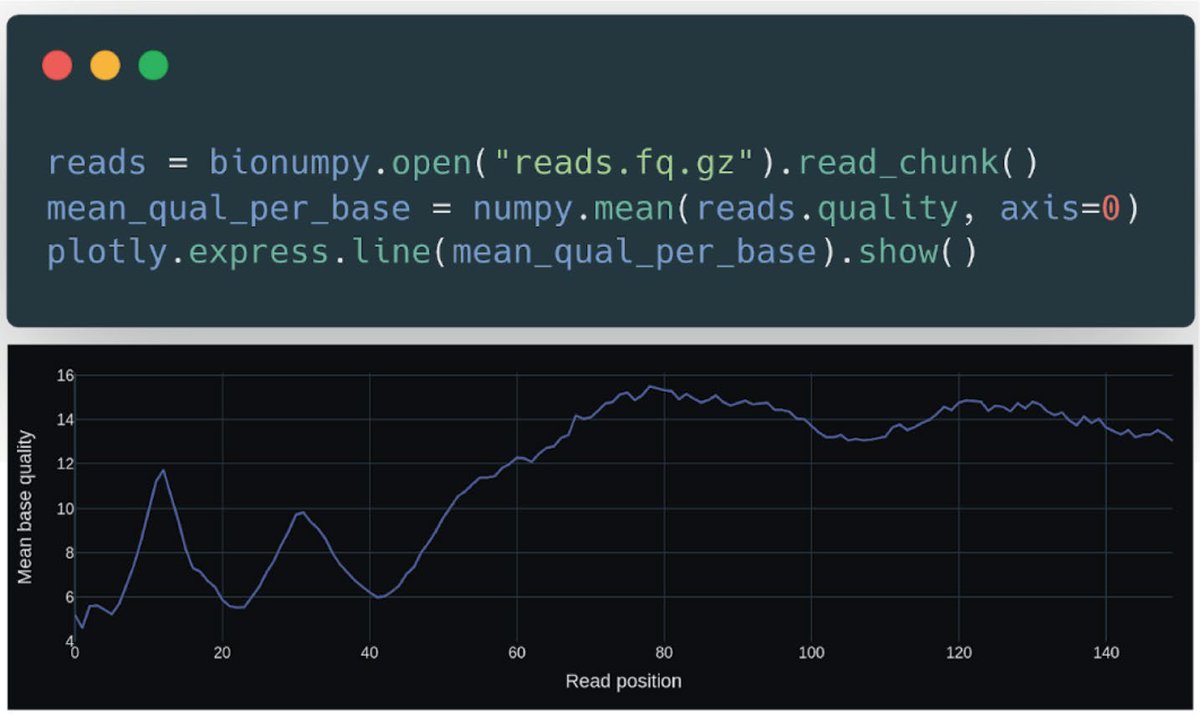
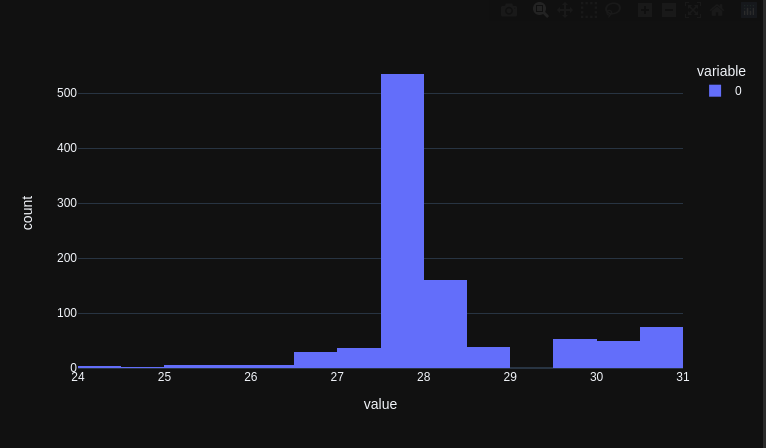
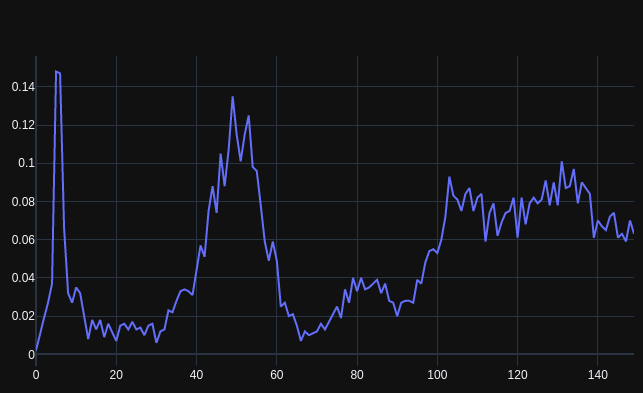
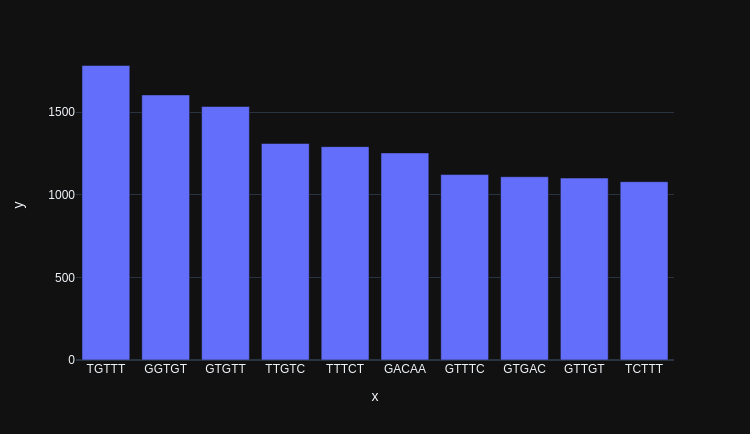
BioNumPy: array programming for biology Nature Methods • BioNumPy revolutionizes biological data analysis by integrating the power of NumPy-like arrays, making Python even more accessible to bioinformaticians. • It enables direct handling of biological formats (like FASTQ,