Michael Stam
@mjstam
Senior ML Scientist @ BioPhoundry | Machine Learning | Protein Design
ID: 1324663325519761410
06-11-2020 10:42:13
84 Tweet
286 Followers
563 Following


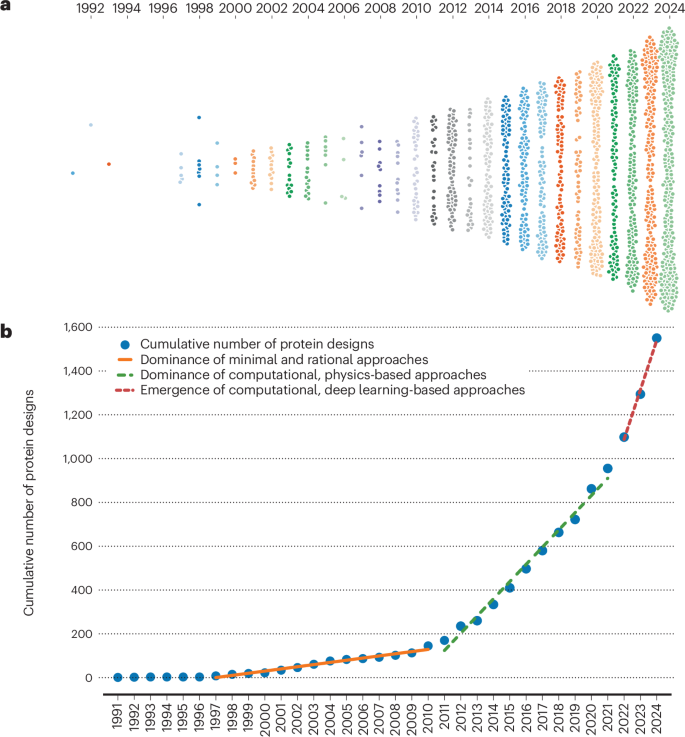
Ever wish there was a way to easily browse designed proteins? You're in luck! Introducing the Protein Design Archive - your go-to resource for exploring designed protein structures across the last 40 years! Website: pragmaticproteindesign.bio.ed.ac.uk/pda/ Preprint: biorxiv.org/content/10.110…


#333: Ft. Elm Town & 🦋 hayleigh.dev, Christopher W. Wood & Michael Stam & Marta Chronowska & WoolfsonLab, Nelson Correia, and Cλris. elmweekly.nl/p/elm-weekly-i… #elmlang



So proud of my lab members applying all our tools to design EGFR binders for the Adaptyv Bio competition. Their designs are currently ranked 30/1000+ from the computational analysis, 🤞 for the binding assay results! Thanks for running the competition Adaptyv Bio! #proteindesign

Excited to announce that our team's designs ranked among the top 30-57 positions in the Adaptyv Bio protein design competition, distinguishing themselves among over 1,000 submissions! So proud of our team! 😀


🎇 We're kicking 2025 off with a Protein Design Archive update, adding structures such as miniaturized electron-transfer proteins, small molecule and protein binders, beta barrels and hairpins, oligomers, and more! Check out all of them at pragmaticproteindesign.bio.ed.ac.uk/pda/ Christopher W. Wood

Our paper on computational design of chemically induced protein interactions is out in nature. Big thanks to all co-authors, especially Anthony Marchand, Stephen Buckley and Bruno Correia nature.com/articles/s4158…

A framework for evaluating how well generative models of protein structure match the distribution of natural structures. Possu Huang Lab


Our paper on the Protein Design Archive has just been published in Nature Biotechnology! It describes our web application for exploring designed proteins (link ⬇️) and contains analysis of how they have changed over time. Marta Chronowska Michael Stam WoolfsonLab nature.com/articles/s4158…