Nils Gehlenborg
@ngehlenborg
Faculty @harvarddbmi. Data Visualization. Biomedical Informatics. Genomics. Epigenomics. Cancer Biology. Single-Cell X. EHR UI. mHealth Data.
ID: 732887081513426946
http://gehlenborglab.org 18-05-2016 10:54:31
250 Tweet
227 Followers
214 Following











An insightful day at our London office yesterday! We were thrilled to host Prof Sir Mene Pangalos FRS, a distinguished member of our Scientific Advisory Board. Our Chief Scientific Officer, Miles Congreve, led a fascinating fireside chat with Sir Mene, diving into crucial topics like selecting the




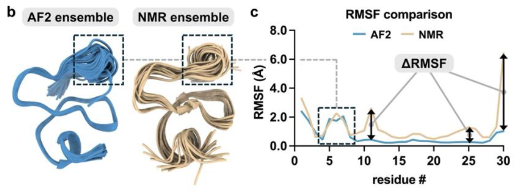
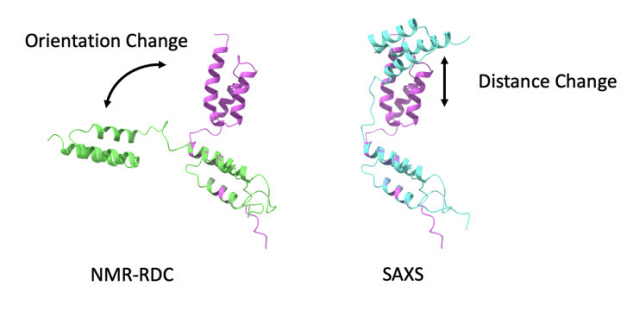
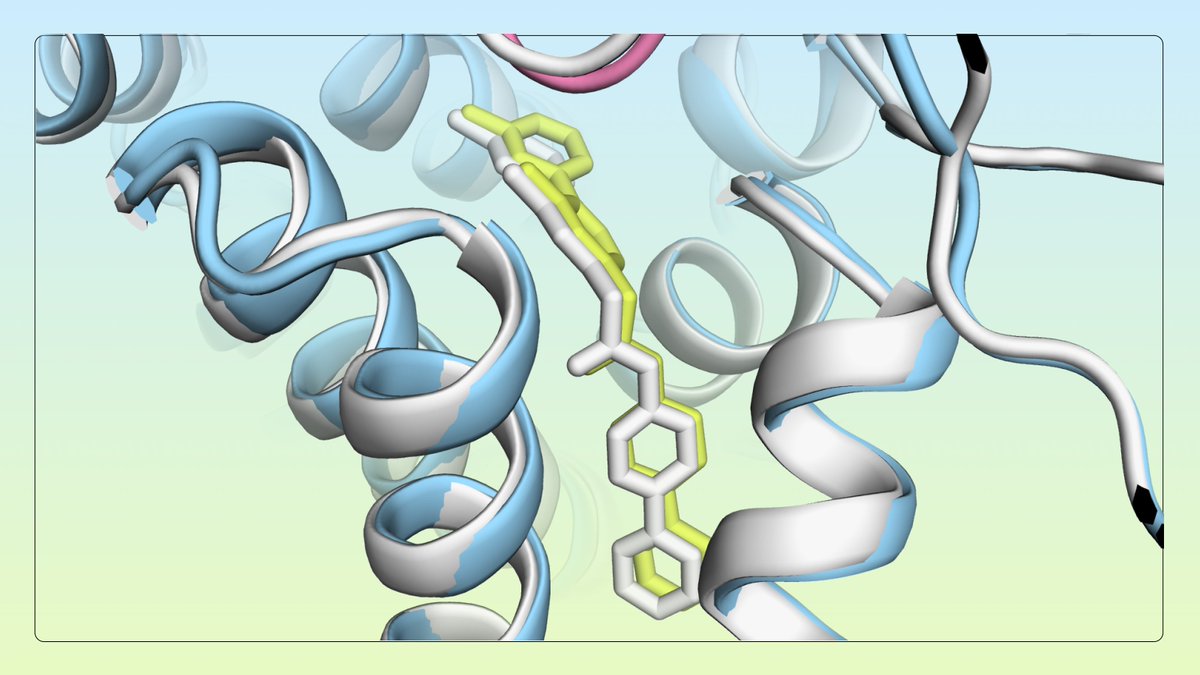
⚕️🧪Delighted to share extended lab validation results for our Google DeepMind Google AI AI co-scientist! Great work from our collaborators Stanford Medicine validating novel insights in liver fibrosis, a condition that affects millions with few effective therapies. More details from