Saro
@pas_saro
AI4Science @MIT • Former Maths @Cambridge_Uni, @AIatMeta, @GRESEARCHjobs
ID: 1266083760317087746
28-05-2020 19:08:10
19 Tweet
409 Followers
150 Following




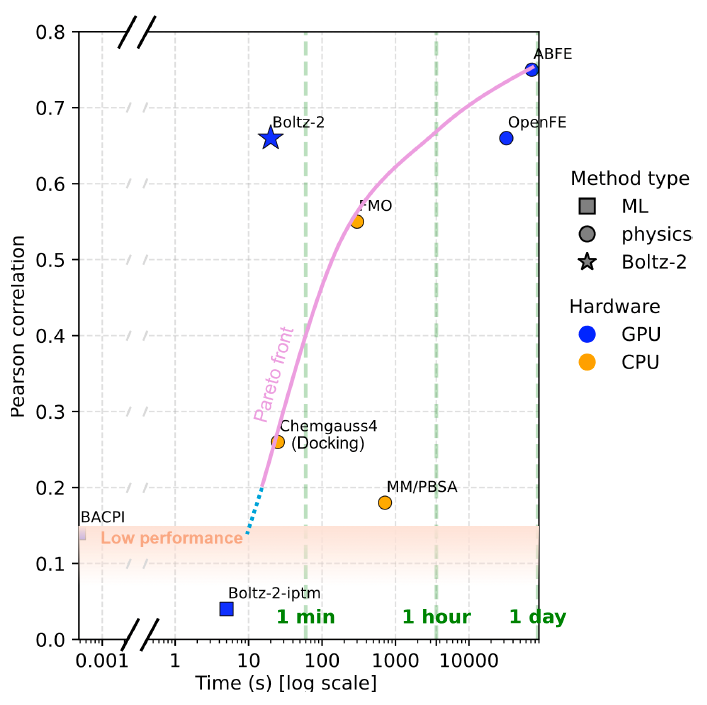
Give this some love - open-source biology AI needs more recognition! huggingface.co/boltz-communit… by Gabriele Corso and team









A team led by Regina Barzilay, a computer science professor at Massachusetts Institute of Technology (MIT), has launched Boltz-2, an algorithm that unites protein folding and prediction of small-molecule binding affinity in one package. cen.acs.org/pharmaceutical…





Open science, activated. Since the release of Boltz-2 last Friday – the new open-source protein structure and protein binding affinity model from Massachusetts Institute of Technology (MIT) and Recursion – we’ve been introducing the model to the broader community and the reception has been terrific. 🔹At #GTCParis