
Ramon Grima
@ramon_grima
Professor of Computational Biology, University of Edinburgh.
Stochastic modelling of gene expression and intracellular biochemical dynamics.
ID: 1453602450787782660
https://grimagroup.bio.ed.ac.uk/ 28-10-2021 06:00:23
76 Tweet
330 Followers
255 Following



#scRNAseq is a multimodal assay producing two count matrices: unspliced & spliced counts (e.g. for RNA velocity). Which should be used for clustering? Can they be used together? If so, how? Tara Chari w/ Gennady Gorin answer some of these questions in biorxiv.org/content/10.110… 1/🧵

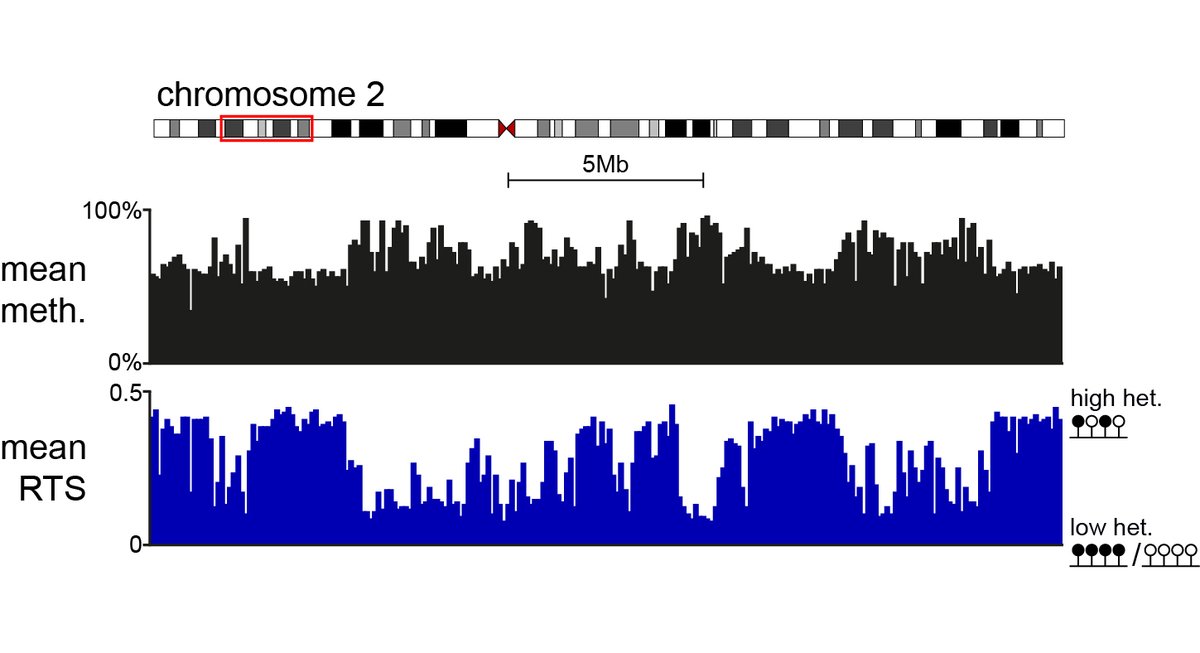
New paper: Genome-wide single-molecule analysis of long-read DNA methylation reveals heterogeneous patterns at heterochromatin that reflect nucleosome organisation MRC_HGU Edinburgh Cancer Research Institute of Genetics and Cancer Biological Sciences | University of Edinburgh Lyndsay Kerr Ioannis Kafetzopoulos Ramon Grima Duncan Sproul doi.org/10.1371/journa…

Job alert📢 We are recruiting for 4 year postdoc fellowships. Up to 3 posts available. If you want to dive into quantitive biomedicine using cutting-edge models and data, this is the place for you. …oss-disciplinary-fellowships.ed.ac.uk ANC@Edinburgh Biological Sciences | University of Edinburgh Centre for Engineering Biology #ai #machinelearning #synbio
Help us understand how DNA methylation patterns are regulated at the single-molecule level using Oxford Nanopore sequencing. PhD available with myself and Ramon Grima MRC_HGU Institute of Genetics and Cancer Biological Sciences | University of Edinburgh, deadline 27th Nov. ed.ac.uk/mrc-human-gene… #epigenetics #Bioinformatics Please RT


Really thrilled to see our work out in the recent issue of Cell! A huge thanks to everyone involved Jan Skotheim Georgi Marinov @lab_reyes William J. Greenleaf Anshul Kundaje (anshulkundaje@bluesky) and the many more not on Twitter. (1/9)
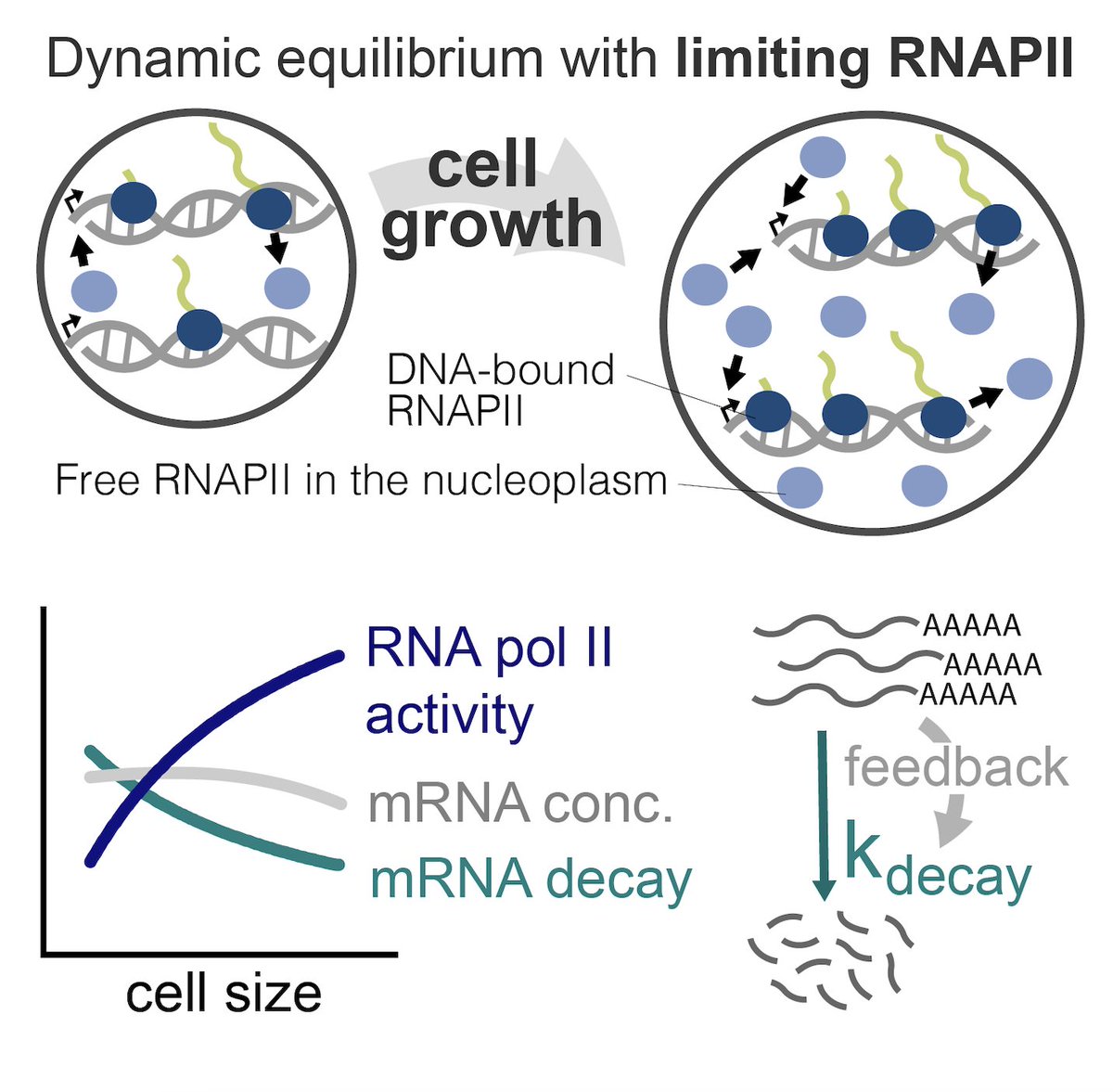


This week's cover features work published in the 2023 Emerging Investigator collection! Impressive modeling of stochastic gene expression by Yiling Wang, Zhenhua Yu, Ramon Grima , & Zhixing Cao (Queen's Chem Eng) Read it here: pubs.aip.org/aip/jcp/articl…


I am thrilled to announce that my new article with Lior Pachter, "New and notable: Revisiting the 'two cultures' through extrinsic noise," is out in Biophysical Journal! cell.com/biophysj/fullt… 1/


How does the bursty character of gene expression vary across the cell-cycle? In recent work with Augustinas Šukys we answer this question by fitting mechanistic models of gene expression to scRNA-seq data in mammalian cells biorxiv.org/content/10.110…


I’m excited to advertise a fully-funded PhD studentship! To find out how you could use maths and stats to understand DNA processes click here: strath.ac.uk/studywithus/po… MathsStats@Strath.


🚨🚨Please RT!!📢📢 Still time to register and submit Poster abstract for bit.ly/EdinPBS2024 ! Higgs Centre for Theoretical Physics Physics Astro @ Edin 💵💷💶We offer Travel Fellowships ! 🚨📢


Excited to share our new paper building a mathematical framework to study developmental tempo differences across species using orbital equivalence 🕐🐁↔️🕑🐒 Grateful to count with Charlotte Manser in the team! journals.biologists.com/dev/article/do…

So excited to announce our new method for multiplexed RNA imaging in bacterial cells: bacterial-MERFISH! A huge congratulations to the team of Ari Sarfatis, Yuanyou Wang, and Nana Twumasi-Ankrah! Check out the following thread or our bioRxiv (biorxiv.org/content/10.110…) 1/12



I have an opening for a postdoctoral research associate in my group at The University of Edinburgh to develop new mathematical models of bursty gene expression and signal transduction and fit these to eukaryotic data. Deadline 24th March. jobs.ac.uk/job/DMA024/pos…


