
Robson F. de Souza
@robfsouza
Biologist/bioinformatician working on sequence analysis at the University of São Paulo
ID: 48724537
19-06-2009 14:49:23
214 Tweet
117 Followers
168 Following

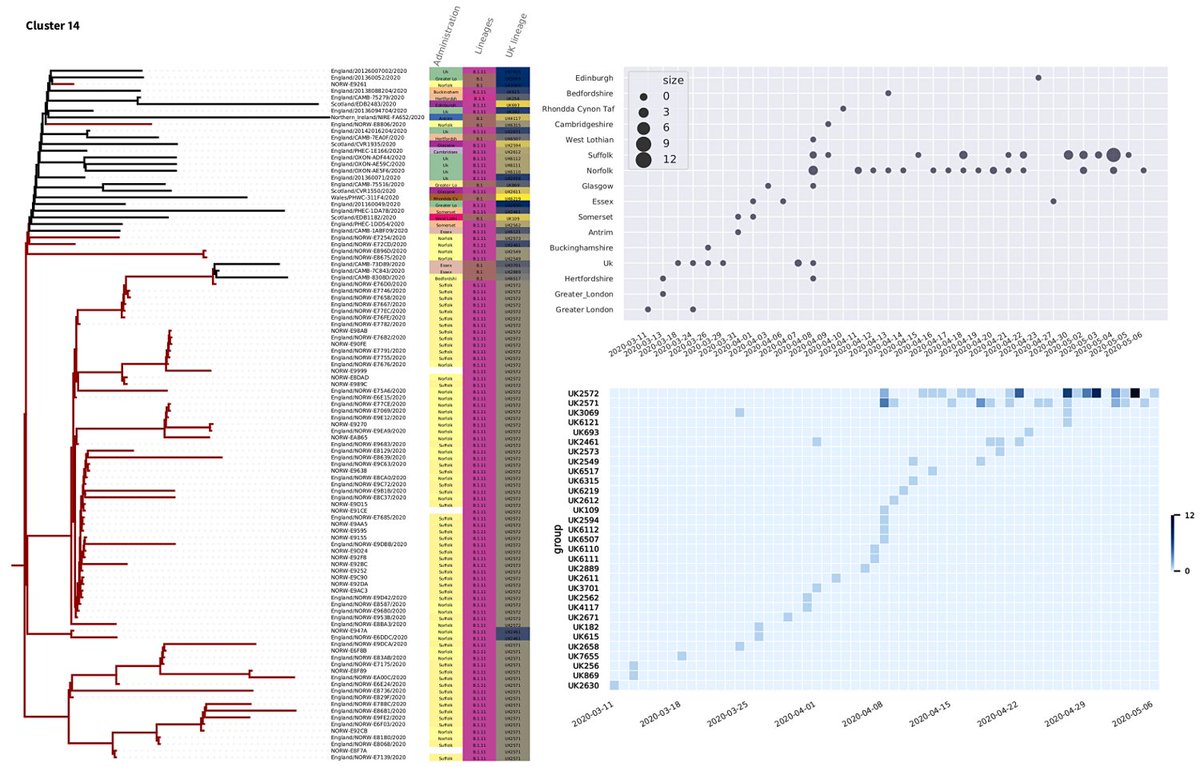
Analysing genomes sequenced Quadram Institute together with the COVID-19 Genomics UK (COG-UK) Consortium database, to inform Norfolk and Norwich University Hospitals clinicians. With a lot of help from many people, behind each dot there's a small army collecting the data, designing the algorithms...











Very proud of my former postdoc mentor, L. Aravind (Aravind group), elected fellow of the American Academy of Microbiology (2022)🔬🦠🧬. One of the most brilliant scientist I've met. Well deserved! 🥂



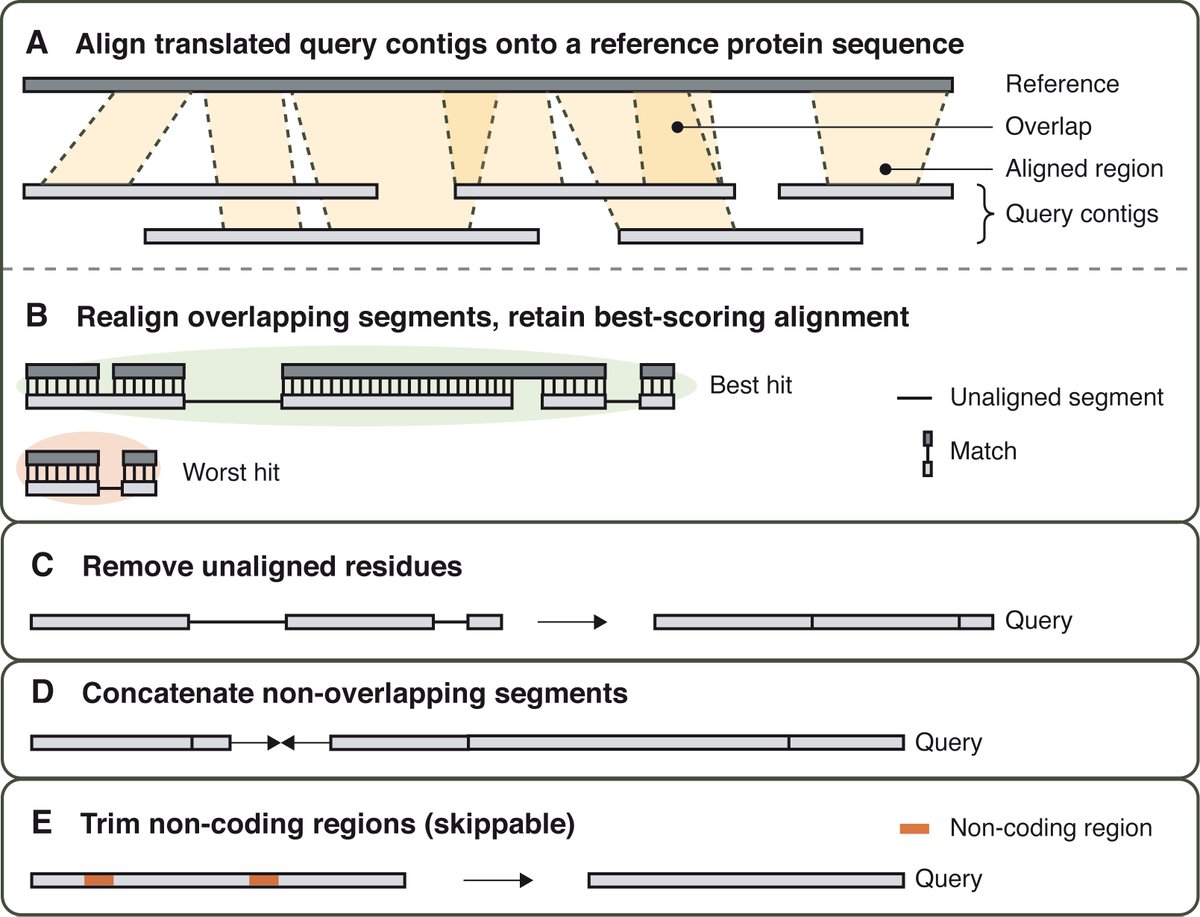
Very happy to see our genome skimming tool Patchwork finally published with GBE (Genome Biology and Evolution)! The software has been developed by @fethalen and Clara Koehne performed the benchmarking 1/2 academic.oup.com/gbe/advance-ar…

In this bioRxiv preprint, we (Kevin Corbett ) present the architecture and activation mechanism of the PARIS antiviral defense system. Thanks to our co-authors, Lisa Liang, Eray Enustun, Ph.D., and Joe Pogliano, for their support in shedding light on this Anti-anti-defense system.🧵


Rejeite a Proposta de Redução do Orçamento da FAPESP! - Assine a petição! chng.it/spLvBqsKQm via Change.org Brasil





