Roberto Rossini
@robomics
Postdoc at UiO developing tools for 3D genomics
ID: 1262367026993078272
18-05-2020 13:01:00
19 Tweet
51 Followers
140 Following

Please RT! Exciting #PhD opportunity to work with Francisco J Ruiz-Ruano Simone Fouché Simone Immler & me at @biouea in beautiful Norwich, to help understand why songbirds have a germline-restricted chromosome. Feel free to DM me for questions. findaphd.com/phds/project/e…


Fully funded PhD position in bioinformatics available in my group. Exciting project focusing on analyzing in-house Hi-C data to model 3D genome alterations in cancer. Please RT. Read more and apply here: jobbnorge.no/en/available-j… Department of Biosciences, University of Oslo International Nucleome Consortium




Very happy to see the peer-reviewed version of this work out at …eneticsandchromatin.biomedcentral.com/articles/10.11… We performed expression-methylation quantitative trait loci in 19 cancer types and identified 13 TFs likely driving DNA methylation patterns around their TFBS in at least 2 cancer types 1/6

Very happy to host Renee Beekman Beekman Lab from Centre for Genomic Regulation (CRG) today for our Furberg seminar. Renée will tell us about "Understanding early lymphoma formation from a single-cell epigenomics and 3D chromatin perspective." Oslo tweeps, join us at 14:30 mn.uio.no/sbi/english/fu…

Out now in GigaScience after substantial improvement since the first version: doi.org/10.1093/gigasc…. Improvements since the first version are briefly summarised in the 🧵

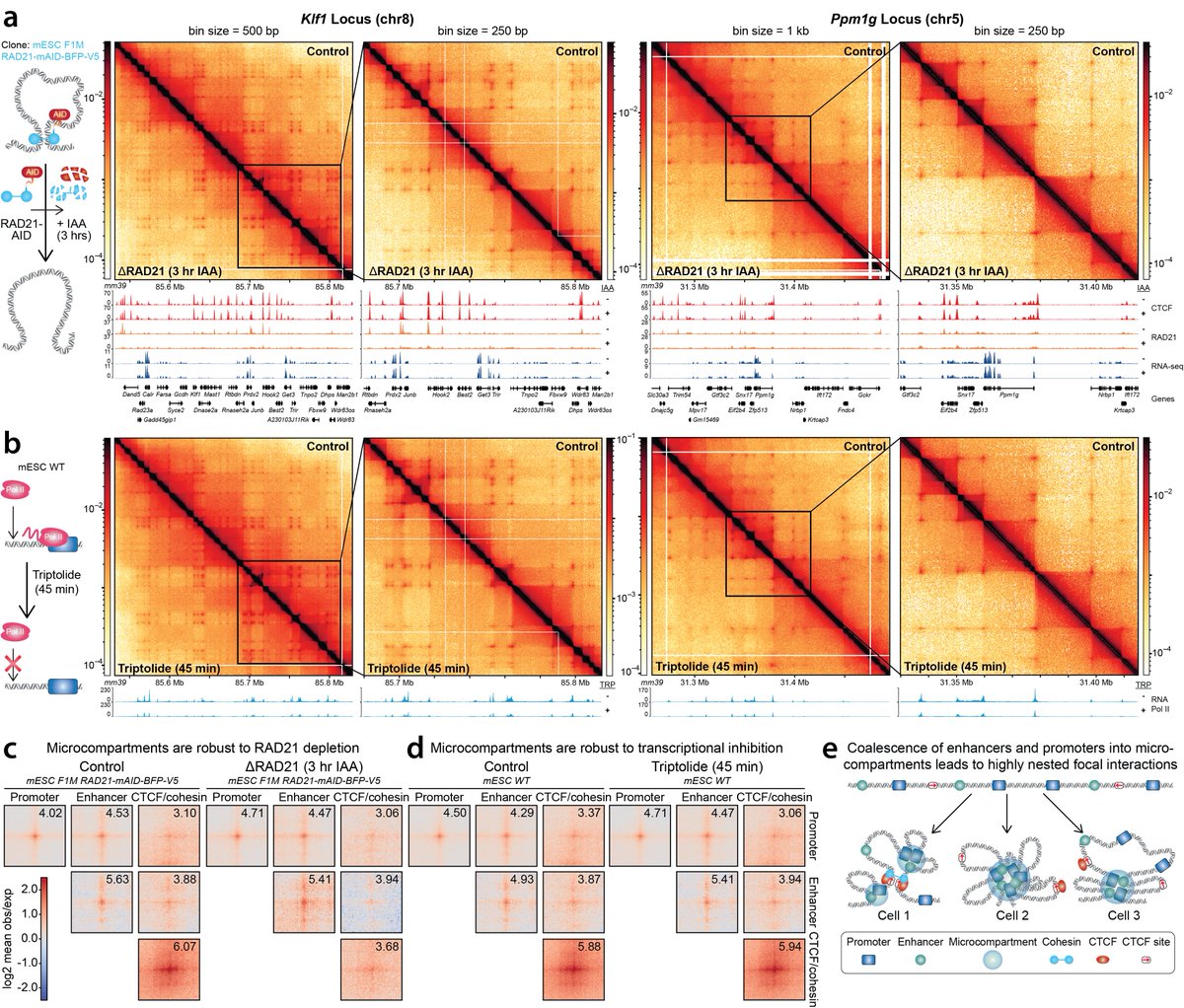
(1/n) Excited to share preprint by Viraat Goel Miles Huseyin We develop Region-Capture Micro-C (RCMC) By focusing on specific regions, we generate the deepest 3D genome maps thus far We see coalescence of enhancer and promoters into “microcompartments” biorxiv.org/content/10.110…

Registration is open for the first Oslo Bioinformatics Workshop Week (Dec 7th-13th) at the University of Oslo. Please see the announcement for schedule, program and registration: mn.uio.no/sbi/english/ev…. Please RT and circulate to interested. RSG-Norway Centre for Bioinformatics, University of Oslo

Great to see the peer-reviewed version of this work now out. Thanks Jonas Paulsen Roberto Rossini for having us involved! And congrats again 👏 genomebiology.biomedcentral.com/articles/10.11…

New publication: "MoDLE: high-performance stochastic modeling of DNA loop extrusion interactions" with Roberto Rossini and Jonas Paulsen Department of Biosciences, University of Oslo. Jonas Paulsen Roberto Rossini Genome Biology genomebiology.biomedcentral.com/articles/10.11…

MoDLE, modeling of DNA loop extrusion, from Roberto Rossini, Jonas Paulsen, simulates DNA contact maps quickly, using low memory. It requires a genome region and the barrier positions in the region eg CTCF binding sites. Predicts effects of deletoins on TADS genomebiology.biomedcentral.com/articles/10.11…



With BioNumPy, I claim my postdocs Ivar Grytten and Knut Rand have given the entire bioinformatics community a Christmas present🫢☺️. This is the fundamental library for working effectively with sequence data that I have been missing since I entered bioinformatics 17 years ago!

A 4-year PhD fellowship in Informatics/Bioinformatics is available at Centre for Bioinformatics, University of Oslo as part of @UiO_LifeSci. The projects will focus on synthetic data generation, applied machine learning and benchmarking within molecular life sciences: jobbnorge.no/en/available-j…. Please RT.


Struggling with Hi-C data? Meet hictk: a new toolkit revolutionizing working with .hic and .cool files! Seamless interoperability for a range of file formats, and blazing-fast performance, 3D genomics has just gotten a lot easier. Developer: Roberto Rossini academic.oup.com/bioinformatics…