Sangsin Lee
@sangsin_lee
Postdoc of @StanfordMed
Muser of @muse
ID: 1242551093365805057
24-03-2020 20:38:36
142 Tweet
167 Followers
1,1K Following





A team led by the Ajo-Franklin group’s very own Biki Kundu has uncovered the mechanism through which E. coli utilizes redox shuttles for extracellular respiration and growth, showcasing the unexpected versatility of microbial metabolism biorxiv.org/cgi/content/sh…

Can AAVs be evolved for better brain delivery after targeted ultrasound insonation? We report new AAVs optimized for focused ultrasound BBB opening. Congratulations to all involved! Richard Hongyi Li Manwal Harb John Heath @JakeSTrippett Mikhail Shapiro (same on bsky) 🇺🇦 nature.com/articles/s4146…

🧬 🧠 Exciting news from Rice BIOE Jerzy Szablowski team! Their innovative gene therapy method could revolutionize brain disorder treatments by targeting specific regions with unprecedented precision. Discover more in Nature Communications! news.rice.edu/news/2024/rese…

Thrilled to announce that I have accepted a TT faculty position at the University of Pittsburgh in Pitt Neurosurgery, starting 01/2025! We will develop noninvasive molecular tools to control and monitor endogenous processes in intact cells and tissues, focusing on the brain. 1/2




I am looking forward to coming back to Massachusetts Institute of Technology (MIT) for Neurotech 2024 tomorrow! I will talk about noninvasive interfacing with the brain, including new projects with our postdoc Sho Watanabe Sho Watanabe and collaborators Vincent Costa and Rui Chen (UCI). neurotech.mit.edu






I am very thankful to BrightFocus Foundation and their donors, for the grant we received today. Using these funds we will enable monitoring of gene expression in the intact retina to understand the molecular basis and progression of age-related macular degeneration and improve diagnosis.



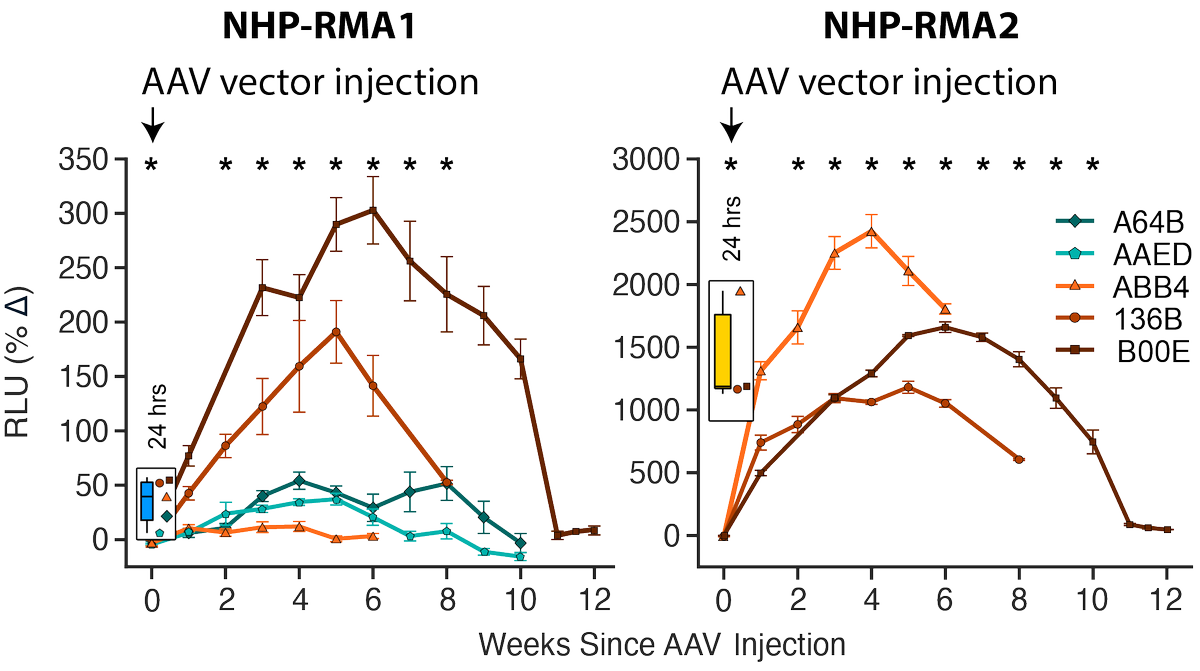
How can you know if gene delivery in a primate worked? Typically through expensive scans or histology. With Vincent Costa and first authors Sangsin Lee and McKenna Romac we made synthetic markers that monitor gene delivery and Cre-dependent expression with a blood test. PDF👇