Saucerman Group
@sauce_lab
Cardiac Systems Biology Group, led by @jsauce7. Tweets by the lab!
ID: 414447526
https://engineering.virginia.edu/faculty/jeffrey-saucerman 17-11-2011 02:13:16
150 Tweet
292 Followers
281 Following

Congratulations Taylor Eggertsen!!! We’re so proud of you!

Go Saucerman Group!!! 💰 💰 💰

Very proud of Taylor Eggertsen for publishing his first paper, "Virtual drug screen reveals context-dependent inhibition of cardiomyocyte hypertrophy"! His network model predicts 52 FDA-approved drugs that regulate heart cell growth. bpspubs.onlinelibrary.wiley.com/doi/abs/10.111…

Saucerman Group’s very own Bryana Nycole Harris has co-founded the Society for Black Biomedical Scientists and Engineers at UVA, along with @Syn_Gibbs! Virginia BME UVA_BIMS 🔬🥽🎓 @DiversityUVASOM UVA Engineering

.UVA School of Medicine PhD students @Syn_Gibbs & Bryana Nycole Harris recently established the Society for Black Biomedical Scientists and Engineers, a platform for Black individuals to unite & thrive within the UVA_BIMS 🔬🥽🎓 & Virginia BME graduate programs UVA. bit.ly/3JtBTGA

Join us for the Virginia BME Emerging Leaders in BME Symposium 9/1! Bryana Nycole Harris Saucerman Group will share her latest on systems biology of cardiac regeneration-development.

Look forward to some great poster presentations this week by Bryana Nycole Harris, Kaitlyn Wintruba, and Diego Trafton from the Saucerman Group! Should be a great BMES meeting! BMES



Come join Saucerman Group to discover drugs for heart failure and regeneration with systems biology! Postdocs UVA CVRC open and free no-GRE PhD app due 12/15 Virginia BME.


Great collaboration led by Shulin and Kyle in McCulloch lab UC San Diego Bioengineering with Philip in my Saucerman Group. We integrated RNAseq and network modeling for a surprise result- the direction of stretch affects the sensitivity of mechanosignaling but not which pathways and genes are sensitive.


Happy to share our preprint, "Logic-based modeling of biological networks with Netflux" by Alex Clark @muktichowkwale and team! Our first paper on Netflux, software that enables biologists and students to model biological networks without programming. biorxiv.org/content/10.110…

Happy to share our latest paper, "Logic-based mechanistic machine learning on high-content images reveals how drugs differentially regulate cardiac fibroblasts" PNASNews, led by Anders Nelson and collab with Kristen Naegle Virginia BME! pnas.org/doi/10.1073/pn…


With machine learning AND human learning, scientists uncover the biological cause and effect of fibroblast behaviors after heart attack. Published in PNAS and featured in UVA Today. at.virginia.edu/AMrne2 Saucerman Group Jeff Saucerman Anders R Nelson


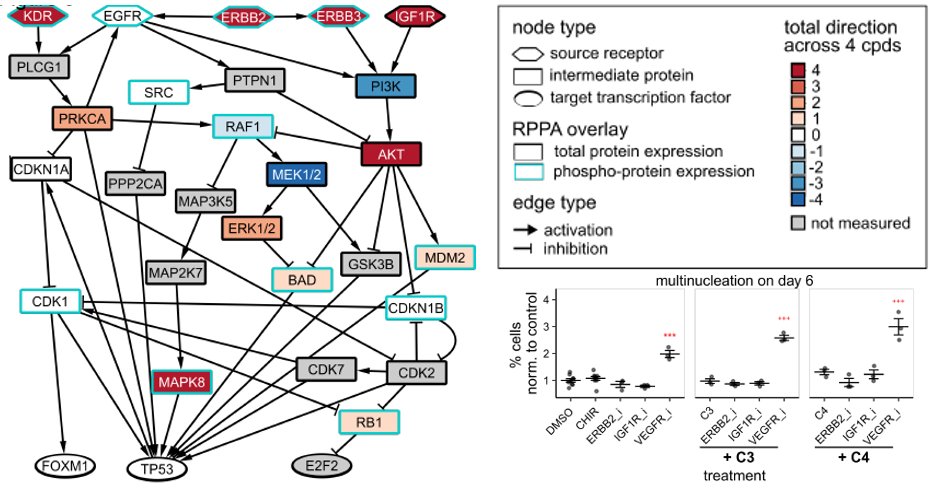
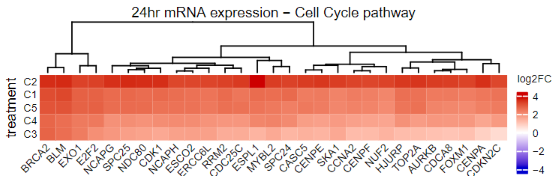
Happy to share the latest paper iScience Int by our interdisciplinary UVA Medicine and AstraZeneca team. We identified conserved mechanisms that explain how small molecules induce proliferation of cardiomyocytes. cell.com/iscience/fullt…
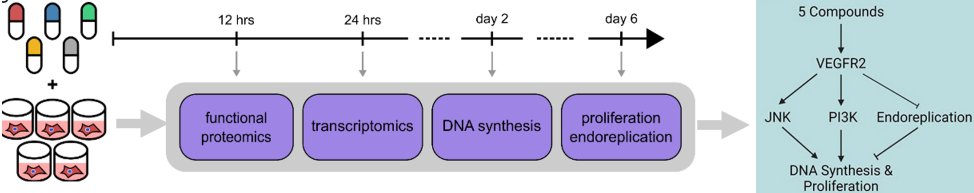


To identify how they work, we profiled human iPSC-derived cardiomyocytes treated with compounds with diverse targets including Wnt pathway, ALK5, CB1R, and COX1/2. We found conserved transcriptomic and proteomic signatures of cardiomyocyte proliferation. 3/ iScience journal


Come meet 6 awesomesauce Saucerman Group lab members AHAMeetings #BCVS2024! Presenting network models and experiments of cardiomyocyte proliferation, hypertrophy, survival, and specification. Virginia BME


Congratulations to Alice Luanpaisanon Saucerman Group on her outstanding Virginia BME PhD proposal! 🎇🫀🎇