Sebastian Deorowicz
@sdeorowicz
Data compression. Algorithms for genome sequencing compresion and analysis.
ID: 1167138749710557184
https://refresh-bio.github.io/ 29-08-2019 18:15:59
125 Tweet
360 Followers
31 Following



Exciting news! 🎉 Our research on ancient phages in the human gut by Piotr is now out in Nature Communications! 📚🔬 A big shoutout to @BEDutilh and Yasas Wijesekara for an amazing collaboration.



After a few years of development, Kmer-db v.2, our tool for finding similar sequences in large collections of genomic data (even millions of viral genomes), is ready. If interested, take a look at the GitHub repo and related paper. github.com/refresh-bio/km… biorxiv.org/content/10.110…







New paper online in Nature Biotechnology by Sebastian Deorowicz group and Salzman Lab: SPLASH2 speeds up analysis of sequence variation in massive datasets.

Happy to share our latest paper with Marek Kokot on SPLASH2 for ultra-efficient reference-free discovery directly on raw sequencing reads out in Nature Biotechnology, supervised by Salzman Lab and Sebastian Deorowicz, and with great contributions from Tavor Baharav. nature.com/articles/s4158…


The latest hifiasm can directly assemble standard Oxford Nanopore simplex R10 reads, without HERRO correction or other preprocessing, to phased contigs of contiguity comparable to HiFi assembly. Like before, you can further add ultra-long, Hi-C or trio data for better assembly.

Recently, our SPLASH paper (nature.com/articles/s4158…) was published in NatBiotech. Now, we release its extended version, sc-SPLASH (biorxiv.org/content/10.110…), which allows reference-free analysis of single-cell data. It was a great experience to work with our collaborators on that!

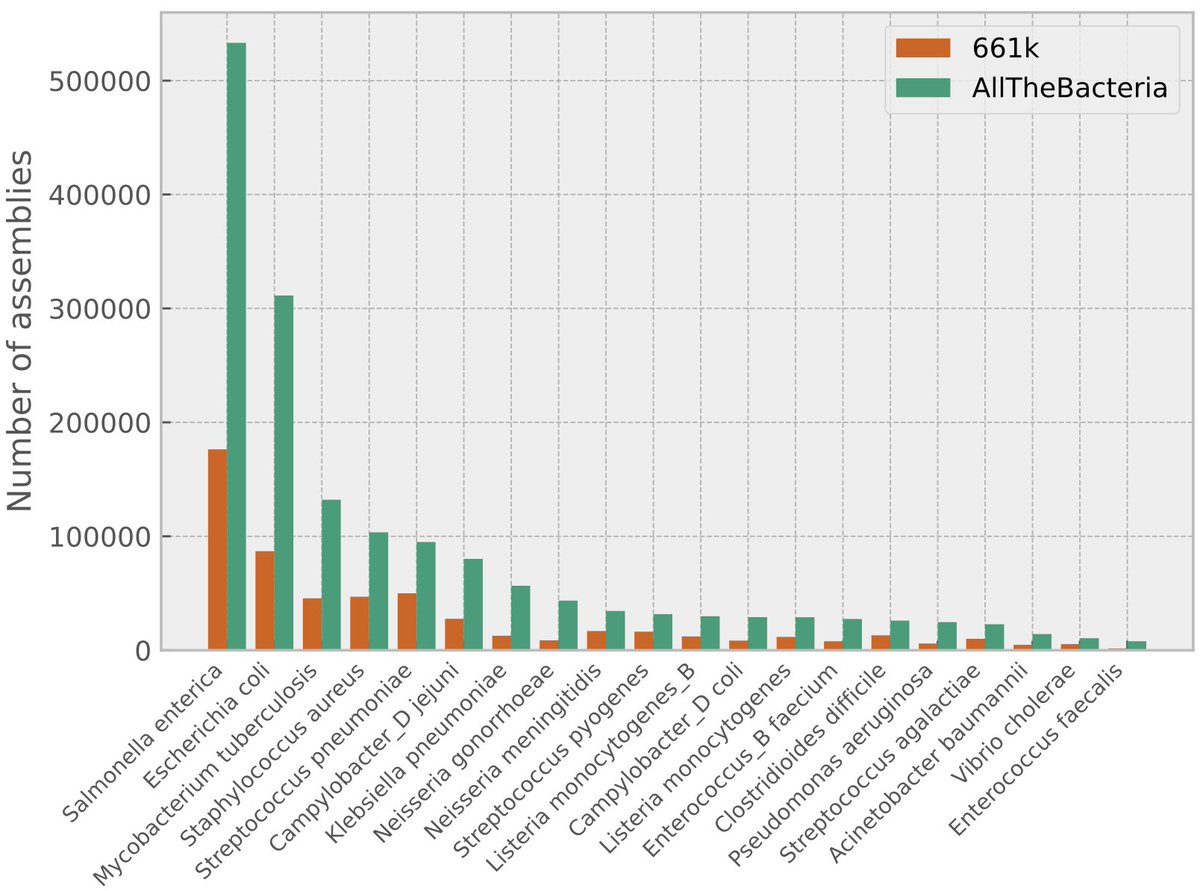
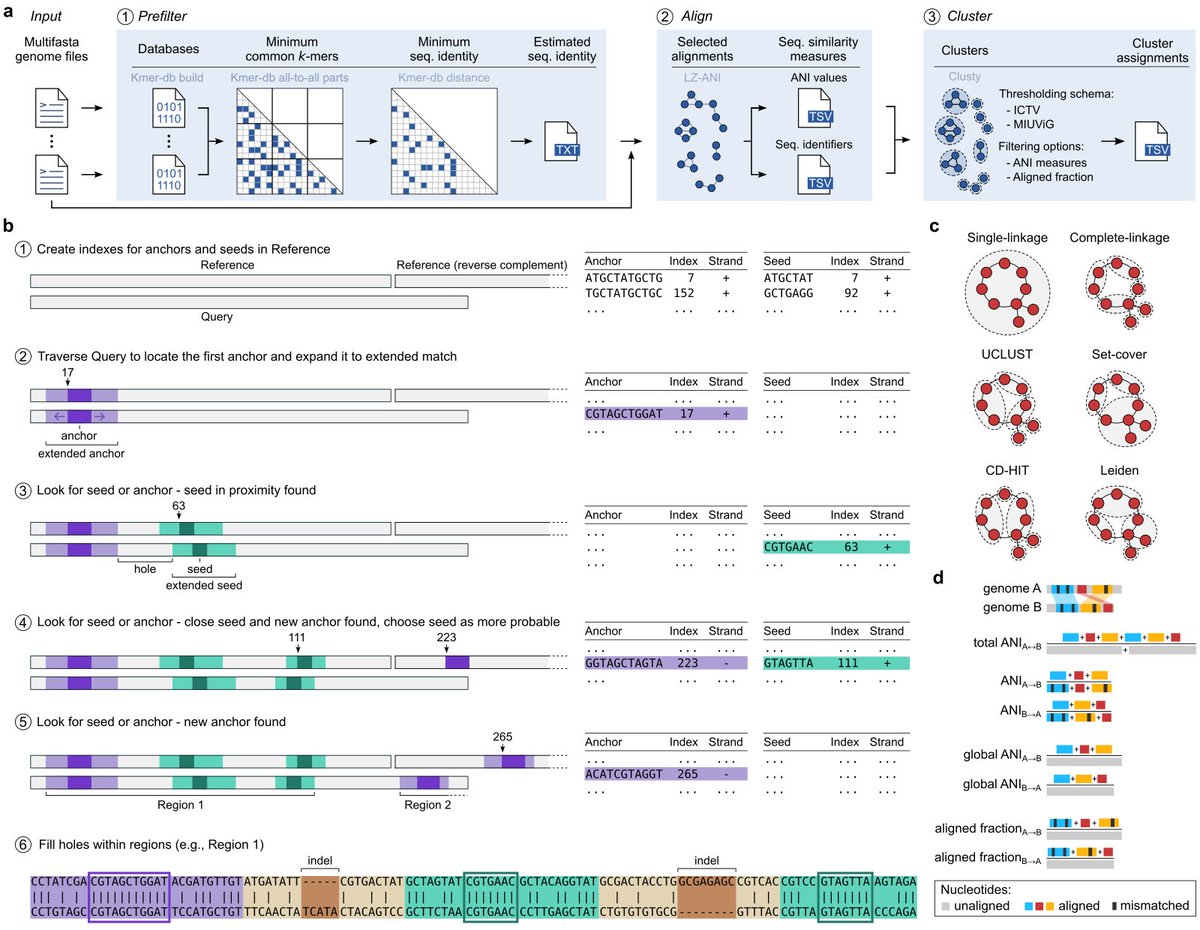
Vclust (the ultra-fast, high-accuracy tool for viral genome comparison & clustering) is now published: nature.com/articles/s4159… Great collaboration with Andrzej Zielezinski, Adam Gudyś, UAM guys, and Bas E.Dutilh

Vclust generates fast and accurate estimation of average nucleotide identity (ANI) for viral genomes, scaling clustering to millions of genomes. Andrzej Zielezinski Adam Gudyś Sebastian Deorowicz Piotr UAM Poznań Politechnika Śląska Universität Jena nature.com/articles/s4159…