
Fritz Sedlazeck
@sedlazeck
Associate Professor
@BCM_HGSC, @RiceCompSci
ID: 2781296741
http://fritzsedlazeck.github.io 23-09-2014 14:13:20
5,5K Tweet
3,3K Followers
390 Following

SPECIAL ISSUE Part 2! This month Genome Research publishes a diverse collection of articles offering novel biological and clinical insights gained using long-read DNA and RNA sequencing technologies and other long molecule approaches. tinyurl.com/Genome-Res-35-4

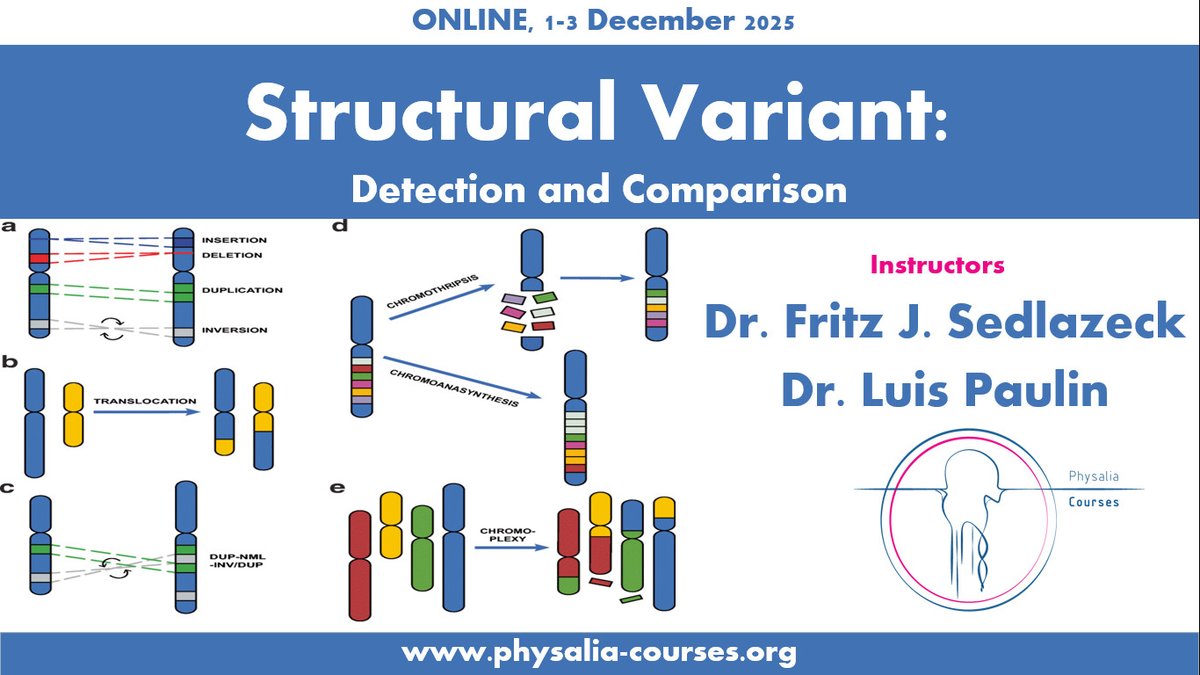
🚨 Course alert! Structural Variant Detection & Comparison with Fritz Sedlazeck & Luis Paulin, PhD 🧬 Learn to identify & analyze germline/somatic SVs using short & long reads Oxford Nanopore PacBio 📅 Dec 1–3 👉physalia-courses.org/courses-worksh…

Come join us at our 7th #bioinformatics #hackathon BCM HGSC From the Labs at Baylor College of Medicine around Structural Variants, Graph genomes & many related topics: fritzsedlazeck.github.io/blog/2025/hack… Groups will work on interesting topics that will be published in F1000. Registration closes 10th of Aug!

1/ 🧬 A Hitchhiker’s Guide to Long-Read Genomic Analysis is out now Genome Research! This mini-review walks through the latest advances in long-read DNA sequencing — from assemblies to variant calling to epigenetics. Link 🔗 genome.cshlp.org/content/35/4/5… 🧵👇


Call for Papers! Genome Research is now encouraging submissions for a Special Issue on The Genetics and Genomics of Somatic Mosaicism, Guest Edited by Drs. Gilad Evrony, Eunjung Alice Lee, and Raheleh Rahbari. tinyurl.com/Call-for-paper… #BOG25 #KSMosaicism25 #MITS25 #somaticmosaicism



Already 30+ registered for our 7th Structural Variants, Graph genomes #bioinformatics #hackathon: fritzsedlazeck.github.io/blog/2025/hack…… Registration closes 10th of Aug! Groups will work on interesting topics that will be published in F1000. BCM HGSC From the Labs at Baylor College of Medicine Rice Computer Science


Do you have an interesting benchmark experiment? We wanna hear about it! Special issue in Genome Biology live now: biomedcentral.com/collections/CO… Submission Deadline: 28 January 2026 #bioinformatic #genetic #genomics please share with your peers!




🧬 Online course: Structural Variant Detection and Comparison with Fritz Sedlazeck & Luis Paulin 🗓 1–3 Dec | Learn to detect, filter & compare SVs from short/long reads Illumina Oxford Nanopore PacBio physalia-courses.org/courses-worksh…

🎉 Congratulations to our Early Career Researcher Pilot and Feasibility Awardees! 👏 Yilei Fu – Fritz Sedlazeck 👏 Cheuk-Ting Law – Kathleen Helen Burns 👏 Yang "Young" Zhang – Zong Lab 👏 Elliot Swanson – Stergachis Lab 👏 Andy Russell – Fei Chen Lab 👏 Hang Su – Broad Institute


Come join us on our SV, graph genome hackathon August 27–29, 2025: fritzsedlazeck.github.io/blog/2025/hack… Seats are filling up! It will be great opportunity to work together on some prototypes. We will publish them together in F1000. Rice University BCMHouston #bioinformatics #openscience

Last call for #bioinformatics #hackathon BCM HGSC Aug 27-29. Register: fritzsedlazeck.github.io/blog/2025/hack… Projects range from TR, SV to pangenomcs, metagenomics etc. Projects will be published in F1000. With support from DNAnexus, Inc. SMaHT Network GREGoR Consortium #openscience #genomics


