Georg Seelig
@seeliglab
ID: 971594970523090944
08-03-2018 03:54:27
1,1K Tweet
1,1K Followers
527 Following






To regulate cellular processes in #syntheticbiology it is useful to bring 2 proteins into proximity, to reconstitute function, guide localization, transcription... We report that from a single 4 helical bundle several pairs can be made. Nature Communications nature.com/articles/s4146… 🧵

Two new posts from Benjamin Maier! One highlights a #preprint Georg Seelig that reports high-throughput sequencing of protein-protein interactions, quantifying more than 100,000 interactions simultaneously. 👉 prelights.biologists.com/highlights/mas… Includes a summary of approaches to study PPIs!

New preprint from our lab and a great collaboration with Georg Seelig ! The anti-cancer compound JTE-607 reveals hidden sequence specificity of the mRNA 3' processing machinery biorxiv.org/content/10.110…

Excited to share our new paper published online at @nature! Congrats to the team, led by super talented students Jordan Kesner and Ziheng Chen nature.com/articles/s4158…



Excellent collaboration with Yongsheng Shi, Liang Liu, Johannes Linder, and Georg Seelig on sequence specificity of the anti-cancer compound JTE-607! Our ML model, Cleavage and Counteraction with Compound 2 on Polyadenylation Outcomes (C3PO), captures polyA sites' JTE-607 sensitivity. 🤖

I am tremendously proud of my University of Washington - MSTP Allen School student Joseph D. Janizek who developed an explainable AI approach for unsupervised gene expression modeling, which led to an exciting collaboration with Matt Kaeberlein's lab on understanding the Alzheimer's disease. genomebiology.biomedcentral.com/articles/10.11…

.Secretary Jennifer Granholm has it wrong. We can’t greenlight the Mountain Valley Pipeline or any new fossil fuel project if we’re serious about moving to renewable energy. The idea that we’ll get to clean energy by tying ourselves to dirty energy for years to come is divorced from reality.



It has been tremendously rewarding to work with outstanding Allen School students, Hugh Chen, Ian Covert, and Scott Lundberg. Our paper that reviews and unifies 26 distinct algorithms to estimate Shapley values is just published in Nature MI! nature.com/articles/s4225…



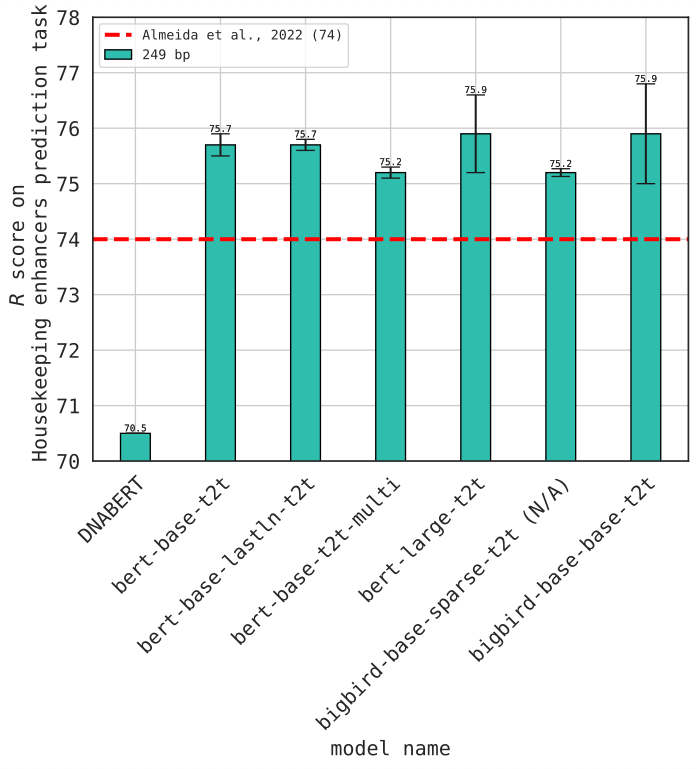
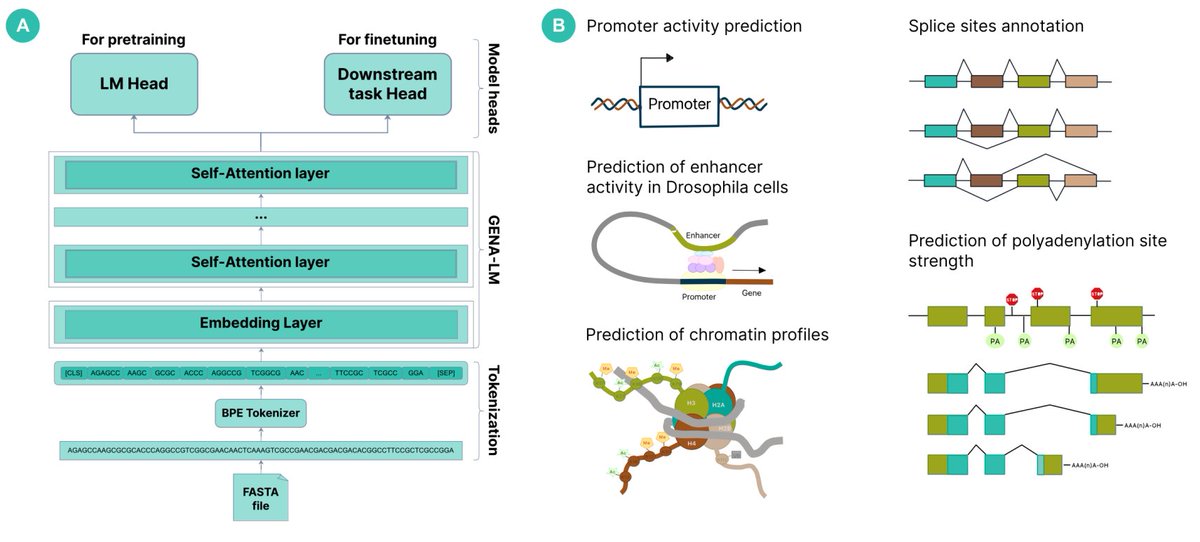
Benchmarks highlight GENA-LM's prowess in inferring diverse biological features from sequence data, from promoters & enhancers (data by Alexander Stark lab) to splice sites & polyA signals (Georg Seelig lab). It performs as well as, and sometimes even exceeds, existing models!5/n