
Joydeb Sinha
@sinha_joydeb
PhD student at Stanford Chemical & Systems Biology. Epigenetics/Mammalian Synthetic Biology @BintuLab
ID: 1204641946066571265
https://scholar.google.com/citations?user=PrHKCOkAAAAJ&hl=en 11-12-2019 06:00:02
53 Tweet
118 Followers
321 Following


Have you ever wondered how to confine bi-stable chromatin states effectively? Then please check out the new preprint from Jan Fabio Nickels and kim sneppen biorxiv.org/content/10.110…


Our latest paper is out today (with A. Xiyal Mukund Josh Tycko and many other talented trainees): High-throughput functional characterization of combinations of transcriptional activators and repressors, at Cell Systems cell.com/cell-systems/f…


We’re excited to announce our new preprint, ‘The H3.3 K36M oncohistone disrupts the establishment of epigenetic memory through loss of DNA methylation,’ expertly led by graduate student Joydeb Sinha! (1/9) biorxiv.org/content/10.110…

It's a double feature on epigenetic memory today! Check another amazing new preprint on single-cell chromatin compaction and memory establishment from wizard postdoc Taihei Fujimori!

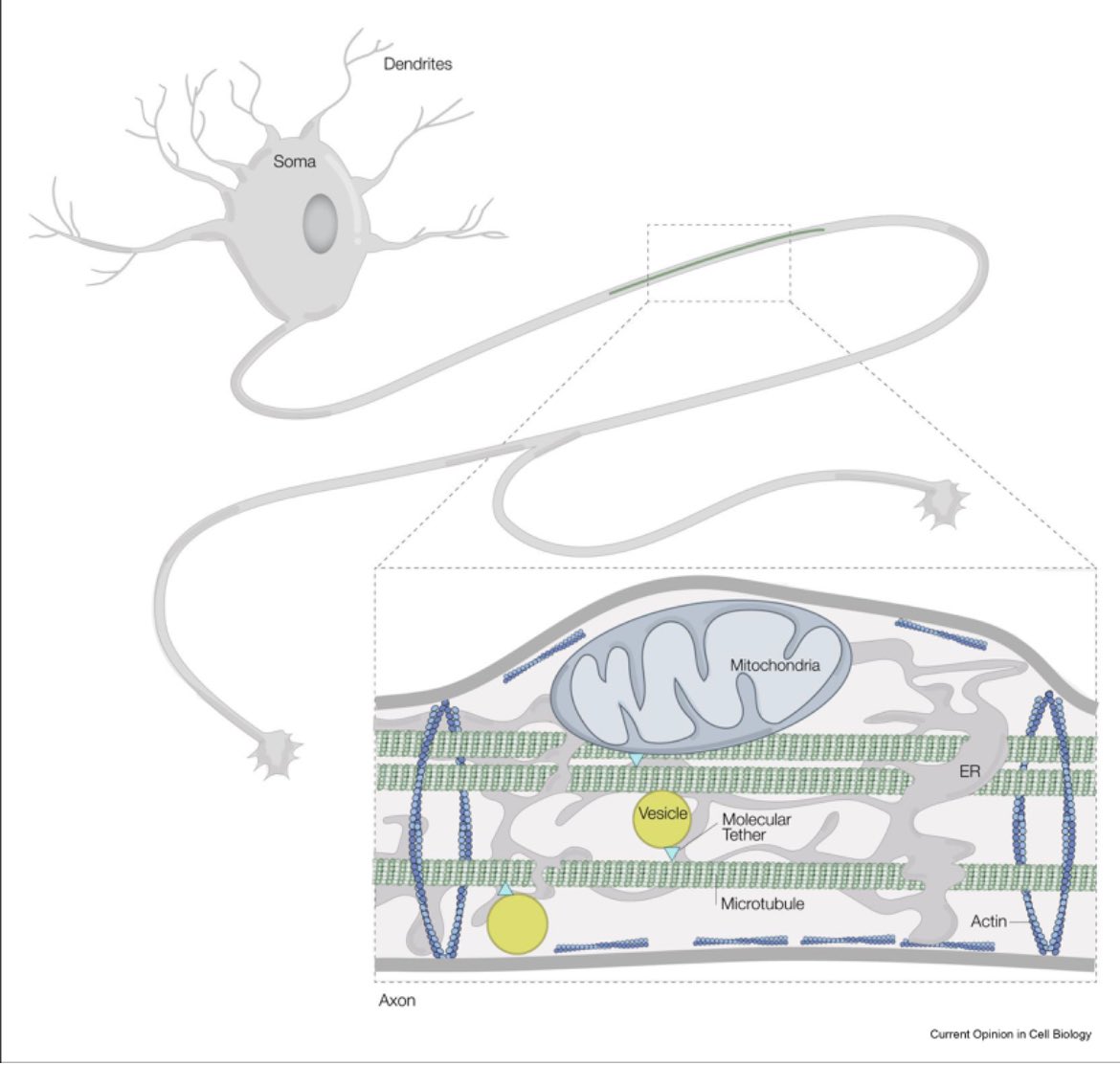
🚨 New paper alert 🚨 I am so happy to share the first paper of my postdoc in Mizuno Lab . Here we hope to summarize the cellular landscape of neurons thru the lens of #cryoET. Hope you enjoy. authors.elsevier.com/c/1ivnv3PA3sYq…




Delighted to see our work tiny.cc/icuazz from the Canzio lab UC San Francisco on how #cohesin-mediated enhancer-promoter interactions shape stochastic #Protocadherin choice profiles out Science Magazine. #epigenetics #chromatin #DNAmethylation #LoopExtrusion #CTCF #3Dgenome. (1/8)




