
Ian Smith
@smithianr
Proteomics Scientist at Velia Therapeutics | Postdoc @jwadeharperlab | Ph.D in @juditvr lab
ID: 1217689718965432325
16-01-2020 06:07:22
68 Tweet
178 Followers
273 Following

We recently shared our biorxiv preprint defining autophagy cargo. With this story Ian Smith created 'CARGO' a web interface to explore all of the data! See if your favorite proteins or organelles are degraded by autophagy during nutrient stress here: harperlab.connect.hms.harvard.edu/CARGO_Cellular…


Are you interested in studying phosphotyrosine modifications on proteins and want to save time and money? Check out our (Judit Villen) automated approach to enrich phosphotyrosine peptides using a pY superbinder or antibodies coupled to magnetic beads! biorxiv.org/content/10.110…

We are delighted to share our work, now available in nature: nature.com/articles/s4158… Our scoring system can be accessed here: kinase-library.phosphosite.org Coming soon --> the tyrosine kinome and an improved system with many new functionalities & features! #TheKinaseLibrary #KL

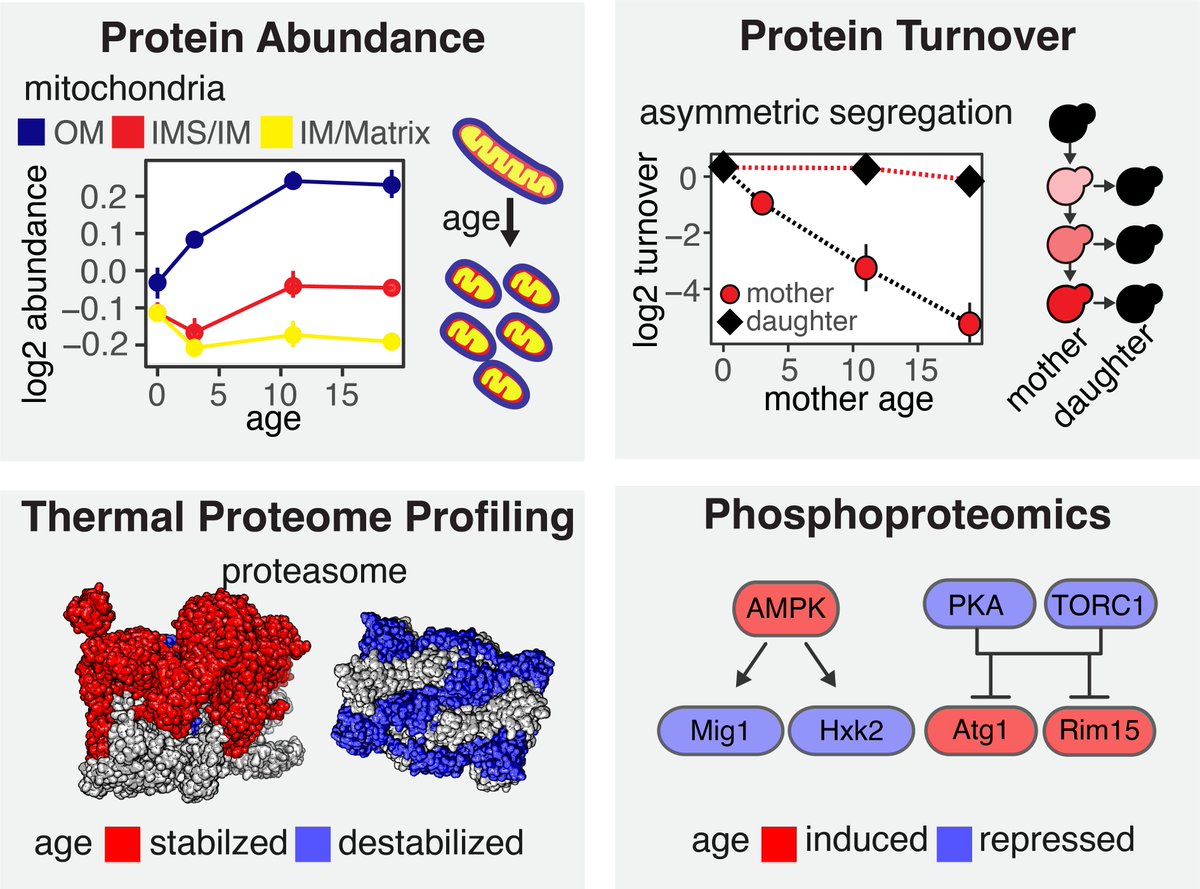
Excited to share our newest #preprint: Multidimensional proteomics identifies molecular trajectories of cellular aging and rejuvenation. A great project with the Judit Villen Maitreya Dunham Matt Kaeberlein teams! 📄Read: biorxiv.org/content/10.110… 💾Explore: rlsproteomics.gs.washington.edu 1/9


We are very happy to announce that our latest story was published in Science Magazine yesterday! 🥳🎉 science.org/doi/10.1126/sc… (1/6)


LOV-Turbo is published today in Nature Methods and plasmids for LOV-Turbo are available on Addgene, here: addgene.org/browse/article… We hope many people will use LOV-Turbo in exciting new ways! nature.com/articles/s4159… If you plan to use NanoLuc BRET to activate LOV-Turbo, please




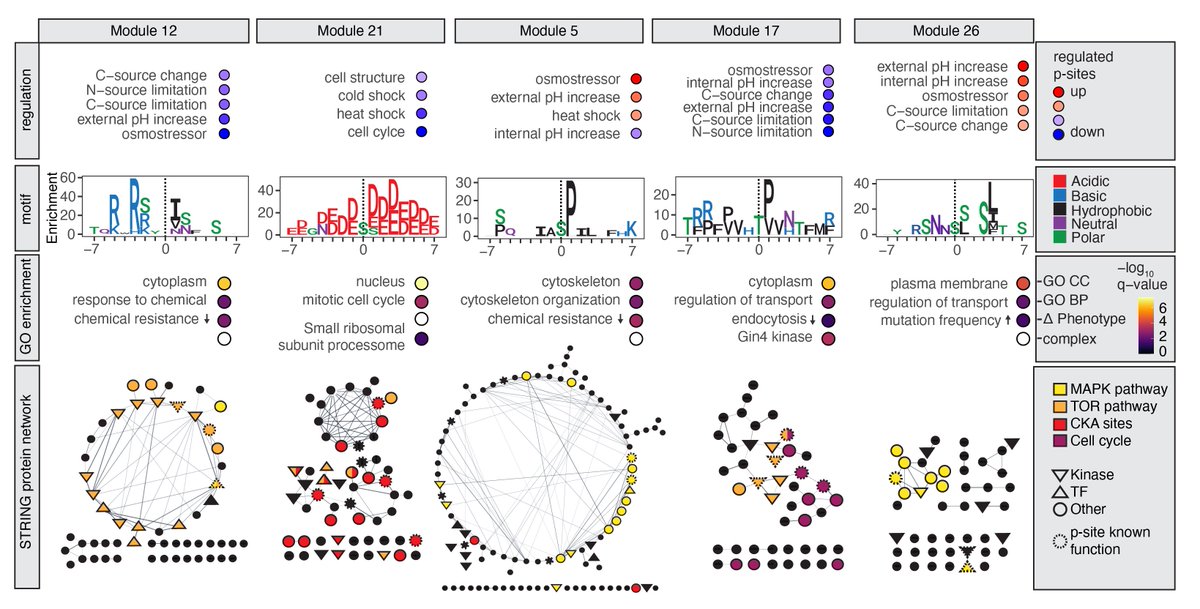
Very happy to share our extensive and systematic Phosphoproteomic Stress Signaling Atlas that is now published NatureStructMolBiol: rdcu.be/doHAL Thanks to the Judit Villen lab and co-autors Anthony Barente , Noelle Fukuda & Ricard RodriguezMias! #proteomics #signaling #TeamMassSpec /1

Thrilled to announce the opening of the Srivatsan Lab (srivatsan-lab.com) at the Fred Hutch Cancer Center. The lab will be building new sequencing technologies to understand how our cells and bodies form over the course of development.

Congratulations Amanda Wiggenhorn, thesis work now in press Nature Communications. Inspired by the structure of the hypothalamic hormone TRH (pyroglu-his-pro-amide), Amanda shows “capping” defines a chemical motif present in many previously unknown bioactive peptides nature.com/articles/s4146…

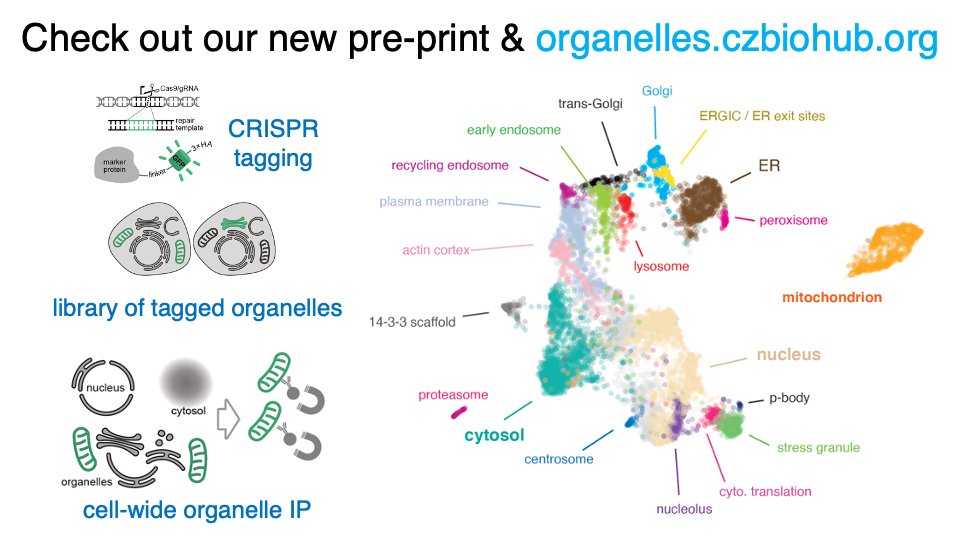
So stoked to be sharing this! New proteomics map of human subcellular architecture. CRISPR + organelle IP mass spec at scale. Check out pre-print biorxiv.org/content/10.110… and organelles.czbiohub.org. 🙌 to fantastic team Chan Zuckerberg Biohub Network SF & collabs! 1/2

We have completed our thorough analysis of ER remodeling during neurogenesis by ER-phagy 🥳@jwadeharperlab Cristina Capitanio Ian Smith Julia Paoli @annabieber_ab Schulman lab @WilflingFlorian. And thanks to the reviewers for their work and insight. nature.com/articles/s4155…


I am excited to share my first project in @jwadeharperlab for feedback. We used spatiotemporal proteomics (1) to describe how cells respond to NLRP3 agonists and (2) to profile the proximal proteome as NLRP3 trafficks to the site of inflammasome nucleation harperlab.pubpub.org/pub/nlrp3/


