Sneha Mitra
@sneha__mitra
Postdoc @MSKCancerCenter Leslie lab
ID: 1126455640841498625
09-05-2019 11:55:28
31 Tweet
148 Followers
303 Following


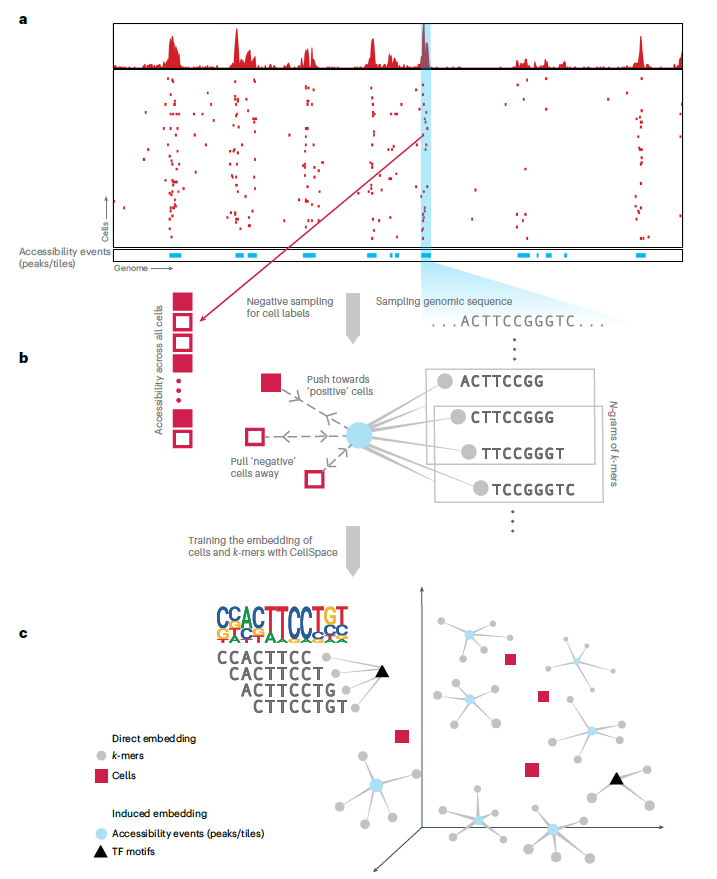
Excited to introduce scKINETICS, an algorithm for joint inference of gene regulatory mechanism and cellular velocity, co-developed by the amazing duo of Cassandra Burdziak and Julia Zhao academic.oup.com/bioinformatics…

🔬🧬 Excited to share our latest study science.org/doi/10.1126/sc… Science Magazine developing ChromExM which uses expansion microscopy to visualize the chromatin, transcription, RNA POL II and pioneer factor Nanog during genome activation.


Excited to share with the single-cell community the most recent work of Tomàs Montserrat Ayuso and AnnaEsteveCodina from CNAG, something we think will be a game-changer in #scRNAseq quality control: biorxiv.org/content/10.110… bioRxiv Get ready for the tweetorial! 1/10



Our work describing #TREM2 #macrophages in #obesity & #diabetes-associated #KidneyDisease is out. An exciting journey since first sighting #TREM2 macs in our kidney #scRNAseq in 2018 w Yiming, Katherine Anna Greka Broad Institute Huge 🙏🙏 to all coauthors dlvr.it/T7FFMb

Introducing pgBoost, by Elizabeth Dorans , using eQTL data to learn consensus variant-gene maps from 10x multiome data. pgBoost is particularly effective in mapping target genes to distal variants where genomic distance starts to underperform. Give it a read!






Out in Nature Communications ! ChromaFold predicts 3D contact maps using single-cell ATAC-seq alone. nature.com/articles/s4146…