Sean Leonard
@spleonard1
Research scientist at a national lab. OIF / OEF veteran. Stutterer. Microbiome engineering, evolution, bees, #BREAD, and data. He/him
ID: 17927372
06-12-2008 19:40:10
13,13K Tweet
2,2K Followers
4,4K Following

I’m excited to announce that Biosphere is coming out of stealth today with $8.8m in Seed funding led by @LowerCarbon & VXI Capital, with support from Founders Fund, GS futures, Caffeinated Capital & B37.




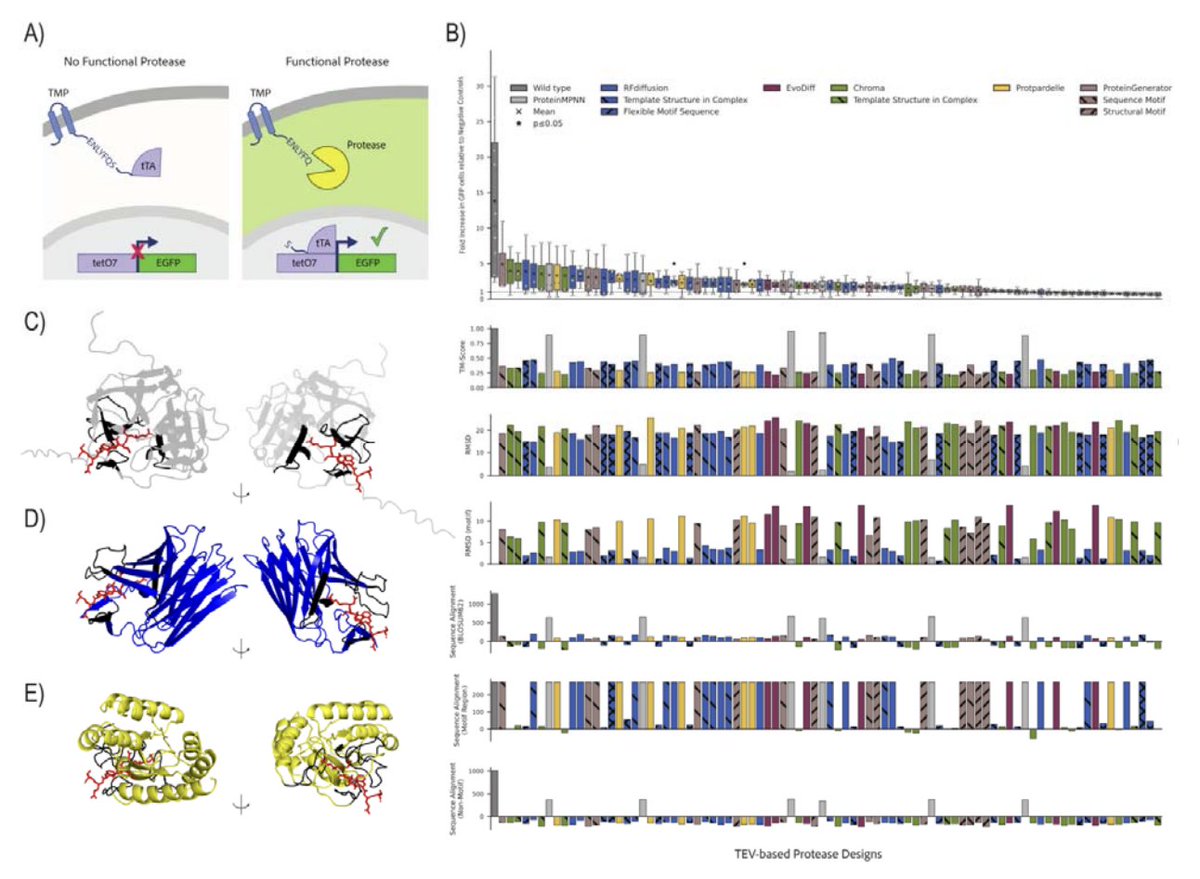
A framework for evaluating how well generative models of protein structure match the distribution of natural structures. Possu Huang Lab














Steered generation for protein optimization: On datasets with ~10^2 measurements, steering a discrete diffusion model outperforms RL on a protein language model for generating improved variants. Jason Yang Wenda Chu Frances Arnold Yisong Yue