Yiyang (Steven) Yu
@stevenyuyy
BME & CS @Columbia | AI x Bio Researcher @ColumbiaMed | Claude Campus Ambassador | Kaggle Competitions Master
ID: 1517177451272622081
http://stevenyuyy.us/ 21-04-2022 16:24:40
52 Tweet
86 Followers
357 Following







We’re excited to introduce Text-to-LoRA: a Hypernetwork that generates task-specific LLM adapters (LoRAs) based on a text description of the task. Catch our presentation at #ICML2025! Paper: arxiv.org/abs/2506.06105 Code: github.com/SakanaAI/Text-… Biological systems are capable of




Happy to introduce AlphaGenome, Google DeepMind's new AI model for genomics. AlphaGenome offers a comprehensive view of the human non-coding genome by predicting the impact of DNA variations. It will deepen our understanding of disease biology and open new avenues of research.

Say hello to the gemini-cli, a local CLI to help you build and maintain software with 1,000 free Gemini 2.5 Pro requests per day : )




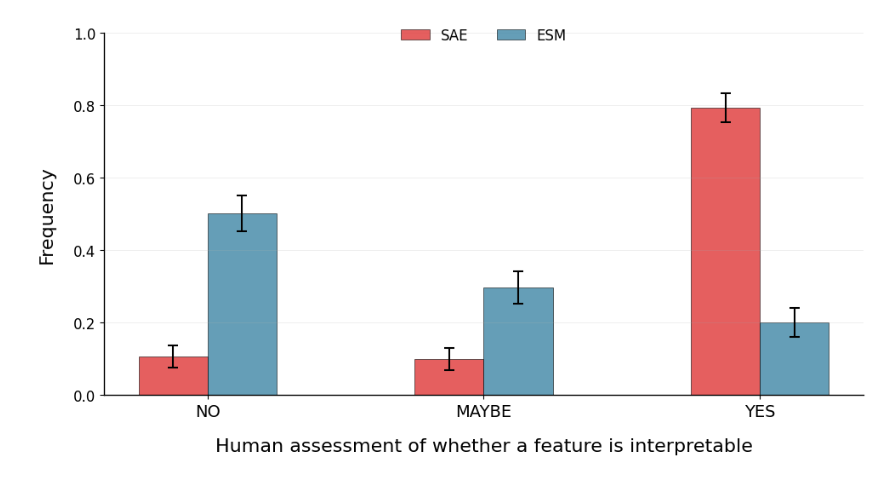
Our work on "Evaluating the representational power of pre-trained DNA language models for regulatory genomics" led by Amber Tang with help from Nirali Somia & Steven Yu is finally published in Genome Biology! Check it out! genomebiology.biomedcentral.com/articles/10.11…