Tailoring Proteins @ DTU Biosustain
@tailor_proteins
X account for Computational Protein Engineering (CPE) research group @DTUBiosustain. PI: Carlos G. Acevedo-Rocha · Hosted by @Lostnorsesailor
ID: 1769643567478177792
https://orbit.dtu.dk/en/organisations/computational-protein-engineering 18-03-2024 08:34:46
44 Tweet
14 Followers
38 Following





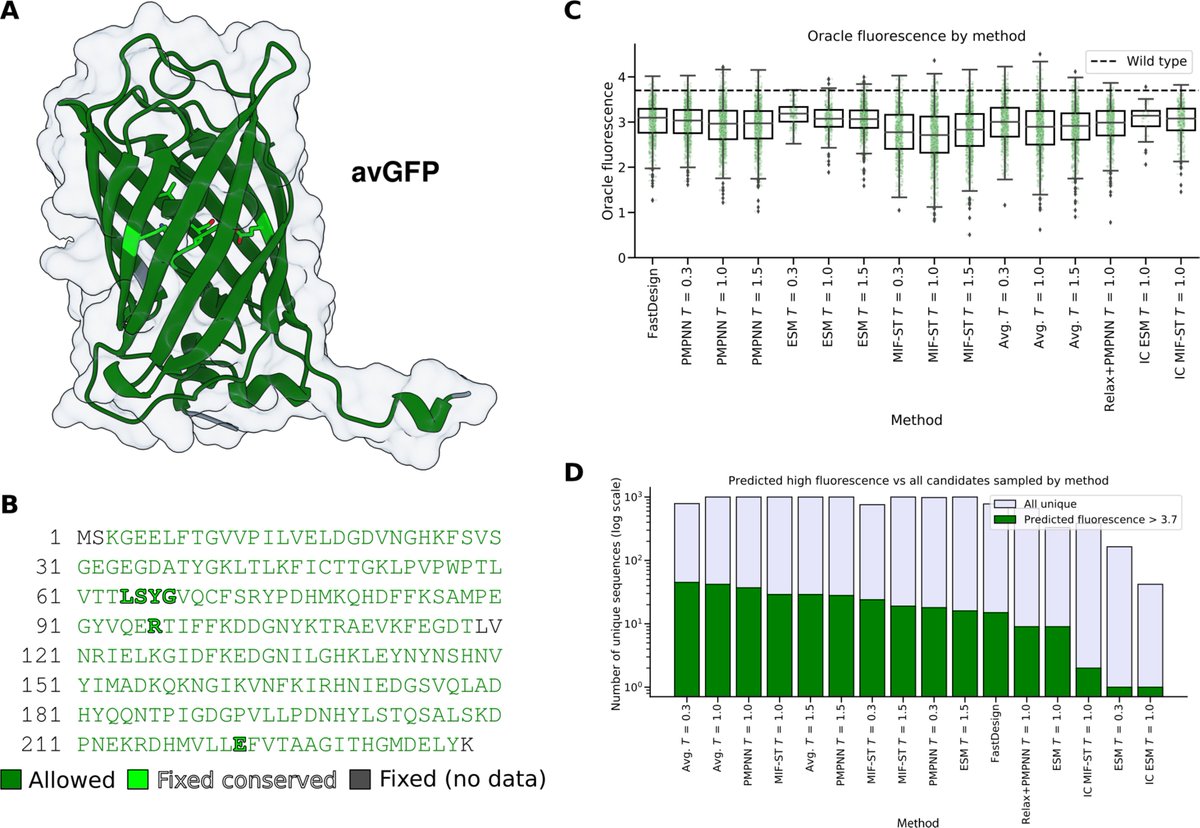
Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates. Bruno Correia Sergey Ovchinnikov biorxiv.org/content/10.110…






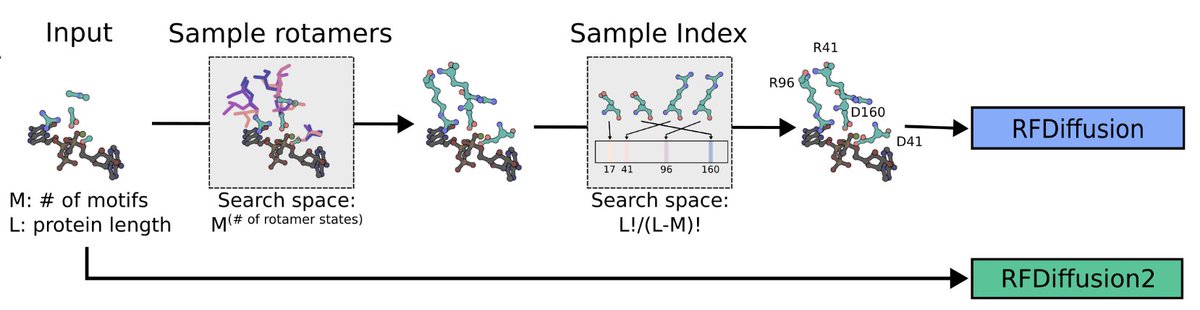
Machine learning-guided directed evolution strategies exceeded or at least matched DE performance with the advantages becoming more pronounced as landscapes had fewer active variants and more local optima. Francesca-Zhoufan Li @ ICLR'25 Yisong Yue Jason Yang Kadina Johnston Frances Arnold