Winston Timp
@timp0

These data provide invaluable information for future work to understand the mechanisms underlying formation and amplification of mitochondrial genome deletions and for using these variants as a metric of aging. Thank you to Winston Timp for collaboration!

So thrilled the Lustgarten Foundation is supporting this cutting-edge project to unlock the inherited genetic basis of pancreatic cancer using long-read sequencing. My co-leaders Michael Schatz Winston Timp are true pioneers in this area. WE ARE HIRING!



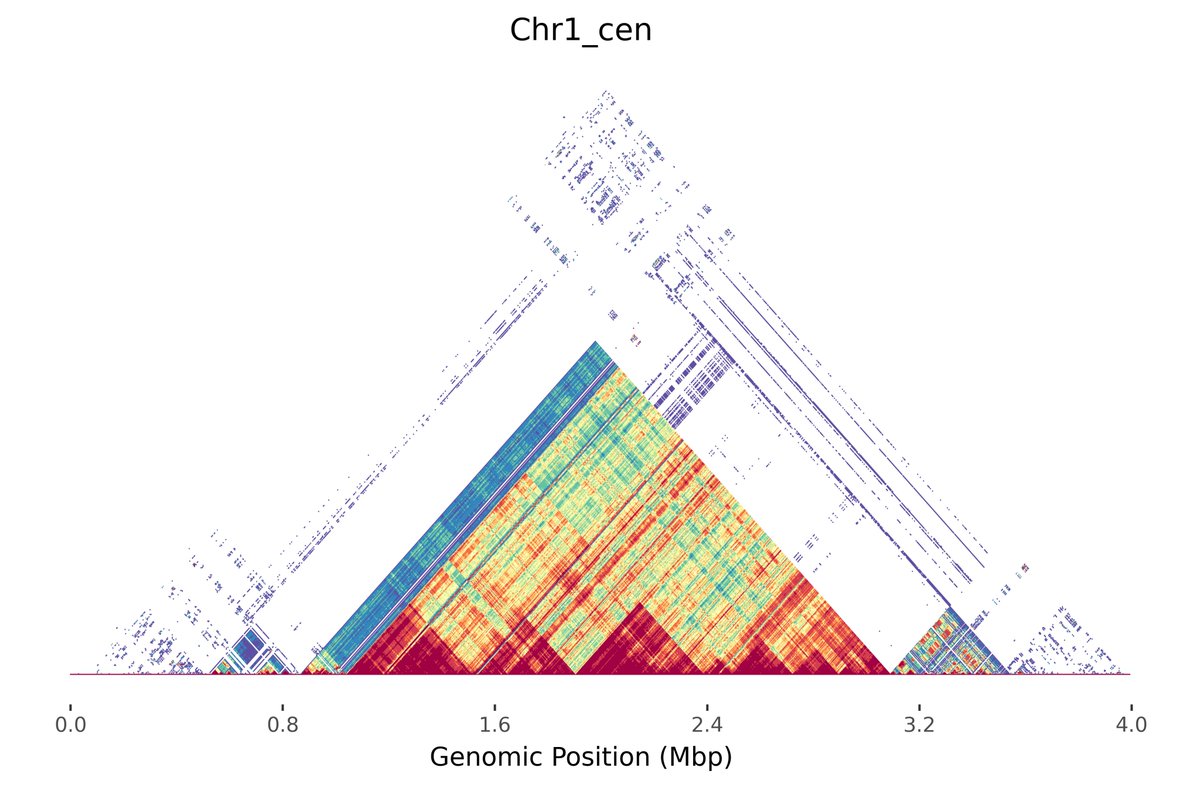
Check out the beta release of Alex Sweeten's new tool Mod.Plot! We "mashed up" Mitchell R. Vollger's StainedGlass visualization with minhashing to enable instant and interactive dotplotting. No alignment needed! Preprint of the method to follow in a couple months 📈

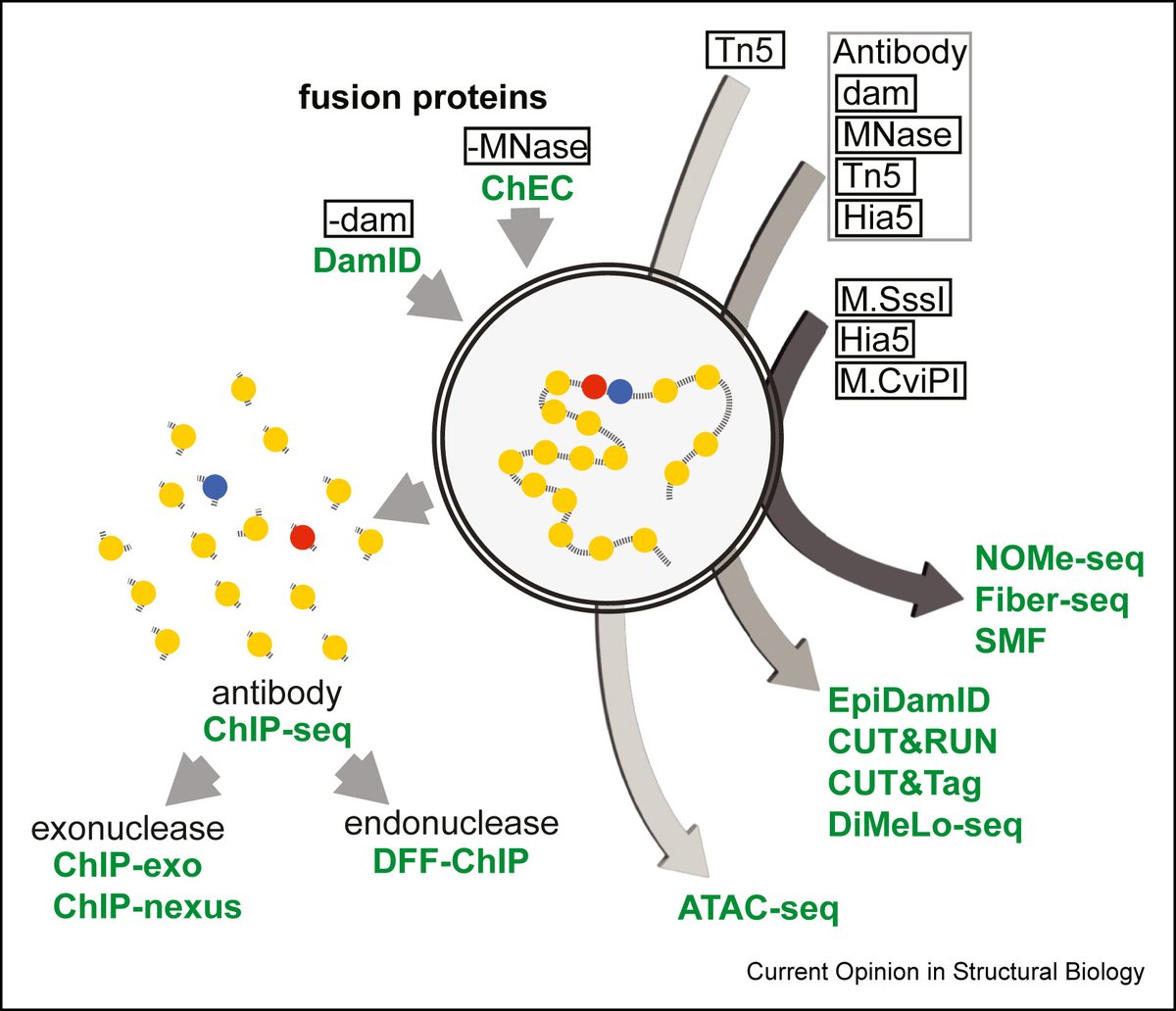
Beyond assembly: the increasing flexibility of single-molecule sequencing technology go.nature.com/3HSiBd4 #Review by Paul W. Hook & Winston Timp Johns Hopkins University Johns Hopkins BME

Our preprint is out! We hacked the Oxford Nanopore sequencer to read amino acids and PTMs along protein strands. This opens up the possibility for barcode sequencing at the protein level for highly multiplexed assays, PTM monitoring, and protein identification! biorxiv.org/content/10.110…





Introducing cornetto, an adaptive genome assembly paradigm using Oxford Nanopore adaptive sampling. - greatly reduces cost per genome assembly - reference agnostic, so works for non-humans - assembly just using saliva - & many more Relies on 2 excellent software #readfish & #hifiasm.


A decade ago, we had thousands of bacterial genomes. Now, we have millions. How to scale computational methods? Our paper in Nature Methods answers this: use evolutionary history to guide compression and search. …From terabytes to tens of GBs… w/@Baym Zamin Iqbal et al. 🧵1/







Direct detection of 8-oxo-dG using nanopore sequencing. #DNAmodifications #Nanopore #Sequencing Oxford Nanopore Nature Communications nature.com/articles/s4146…