Mattia Toninelli
@toninells
Computational Biology PhD Student at @LabPagani w/ @semm_it and bicycle/running junkie.
ID: 1260700671675895808
13-05-2020 22:37:28
177 Tweet
93 Followers
401 Following



Virtual cell models have the potential to transform biological research. Today Chan Zuckerberg Initiative released an initial set of models, including scGenePT and SubCell, designed to be easy to run and build upon. This is a really exciting time in biology. czi.co/4g1jTSa







Our work on 3D genome organization in human cancer is out! In collaboration with Yanding Zhao Howard Chang William J. Greenleaf and TCGA we used HiChIP to profile enhancer connectivity and rewiring in primary tumors. nature.com/articles/s4158…


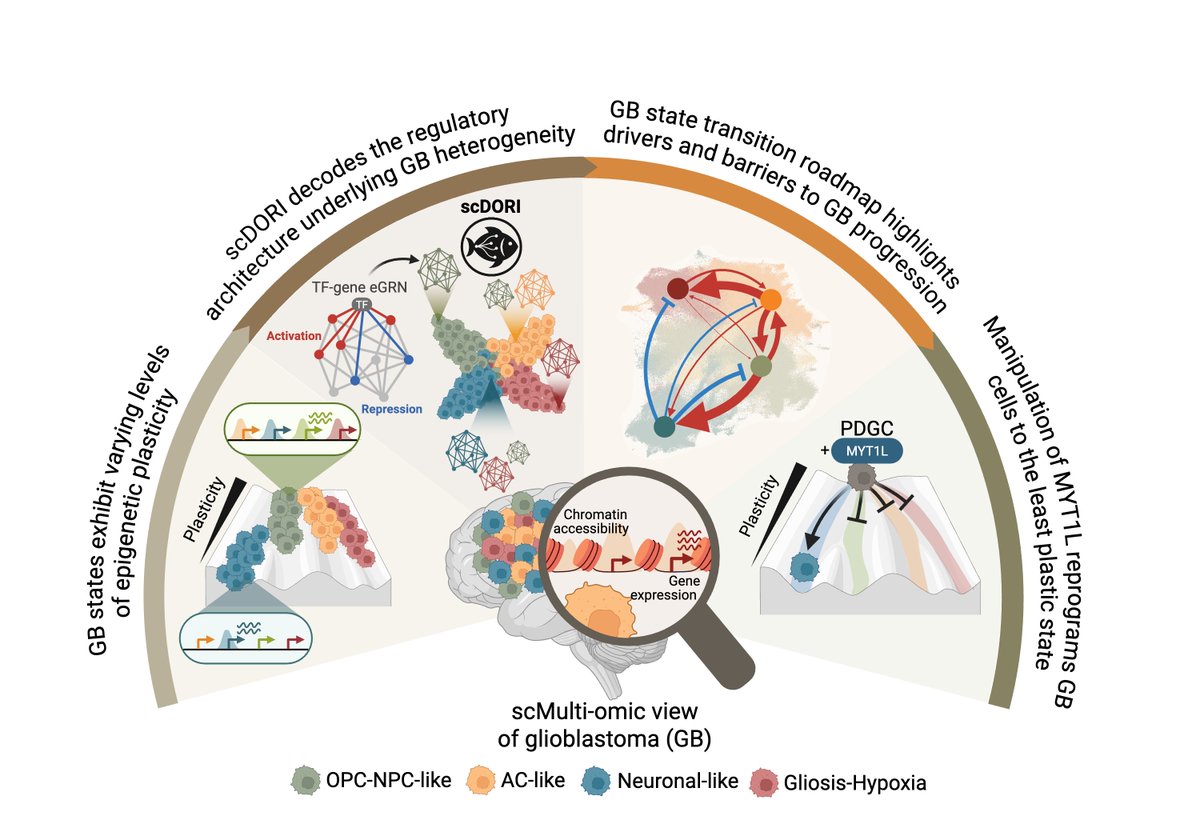
🧠 Excited to share my main PhD project! We mapped the regulatory rules governing Glioblastoma plasticity using single-cell multi-omics and deep learning. This work is part of a two-paper series with Omer Ali Bayraktar , Oliver Stegle and Moritz Mall groups. Preprint at end🧵👇




Amazing days at #EACR2025! Inspiring science, excellent sessions, and such an incredible #cancerResearch community to be part of✨ huge thanks to Cereda Lab and Matteo Cereda for the constant support and IIGM . Also super happy that Fondazione AIRC per la ricerca sul cancro stopped by my poster 🌟