Uday Evani
@udayevani
Interests: Genomics and Bioinformatics
ID: 41651194
http://scholar.google.com/citations?user=5_zHw-IAAAAJ&hl=en&oi=ao 21-05-2009 18:59:29
824 Tweet
181 Followers
580 Following

edyeet is a fork of Chirag Jain's mashmap2 focused on obtaining base-level alignment using edlib. It's faster than minimap2 for variation graph induction, sensitive to repeats, and provides mashmap's probabilistic guarantees of alignment length and quality github.com/ekg/edyeet


Clear Demonstration of Reinforcement Learning Relevant to #AI and #MachineLearning Students by - #BigData #ML #ArtificialIntelligence #DeepLearning #MI #DataScience #DL #MachineIntelligence Cc: Dr Maggie Lieu Mike Gualtieri Jennifer Stirrup #MBA Topics: #AI #Data #Strategy Shannon Platz

Newest results from me and all my colleagues at Deciphering Developmental Disorders Wellcome Sanger Institute. We designed a new tool (InDelible) that uses split reads from exome sequencing to identify breakpoints of structural variants missed by other approaches: medrxiv.org/content/10.110…






My team NY Genome Center is growing. Looking for a highly motivated, detail-oriented Bioinformatics Programmer to help us develop novel tools for whole-genome sequencing data analysis. Please spread the word: nygenome-openhire.silkroad.com/epostings/inde…

If you could read the DNA of half a million people, would you do it and why? Would it work? What would you learn? To answer this and several related questions, let me walk you through the work my Regeneron Genetics Center colleagues just published in Nature. 1/24 nature.com/articles/s4158…



I am excited about our (Genome in a Bottle Jason Chin Heng Li &many more) paper today in Nature Biotechnology to characterize 395 challenging medically relevant genes& errors in GRCH37&GRCH38 that impact analysis nature.com/articles/s4158… BCM HGSC #Bioinformatics DNAnexus, Inc. From the Labs at Baylor College of Medicine



Very excited to see our paper on the high-coverage sequencing and analysis of the samples from the 1000 Genomes project come out. It was a wonderful collaboration with Marta Byrska-Bishop @AniaOkulaBasile and Xuefang Zhao from Talkowski Lab. Please check out this awesome thread!


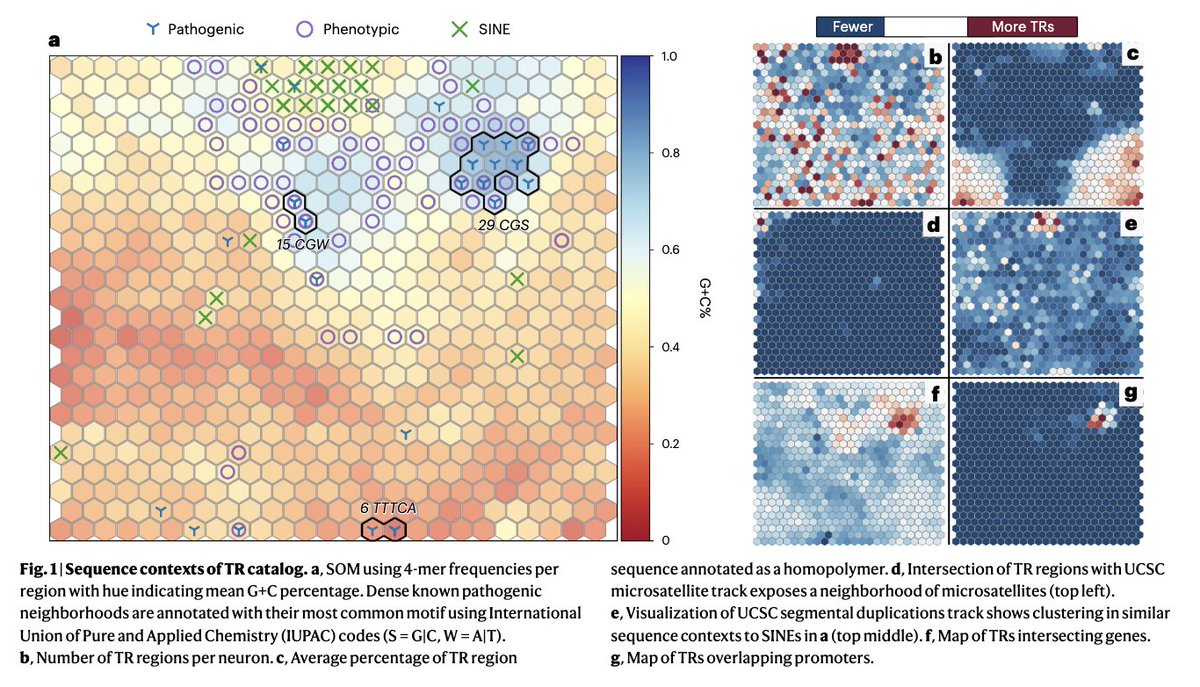
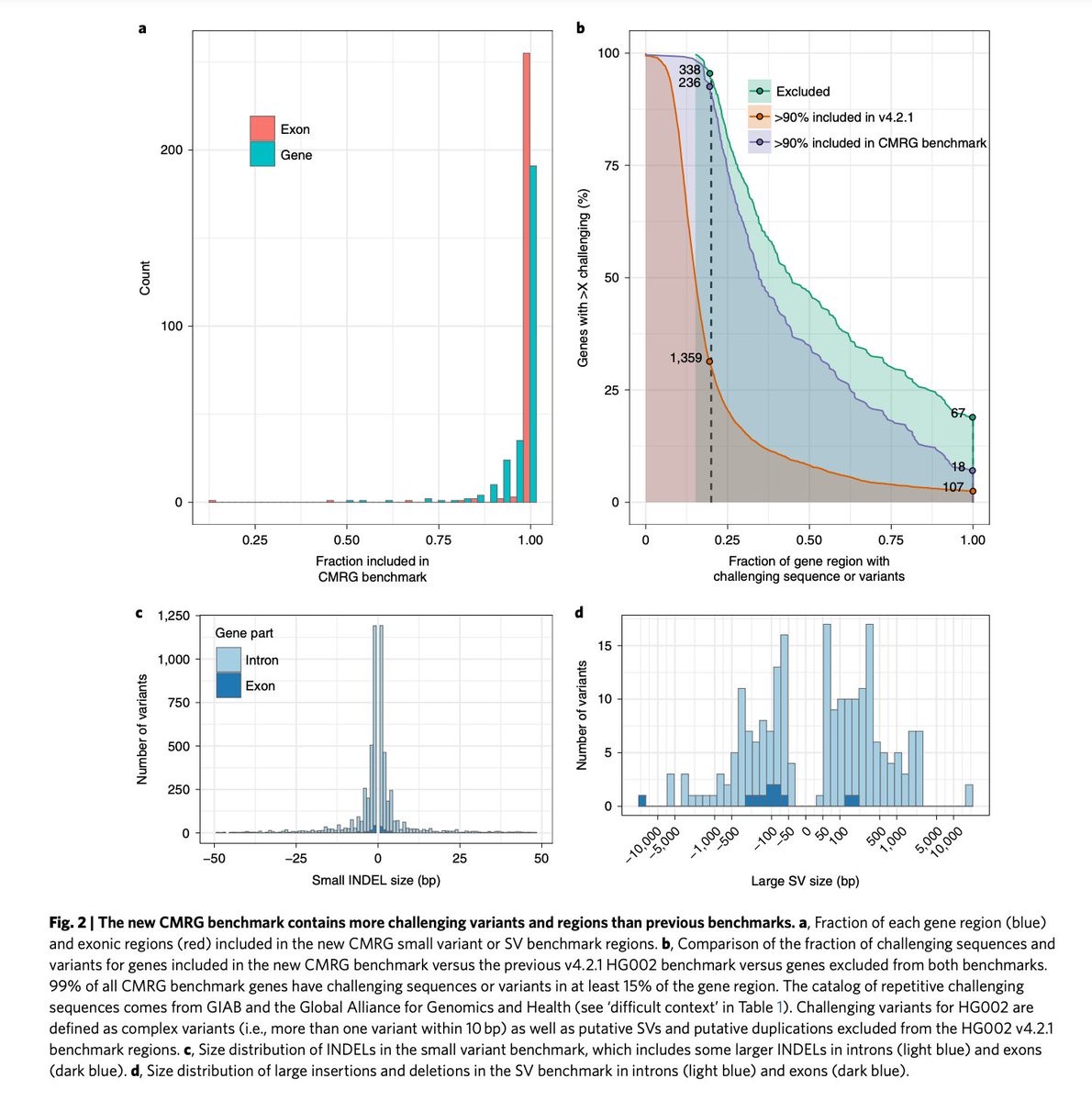
Tandem repeat Genome in a Bottle benchmark Nature Biotechnology out today: rdcu.be/dFQNN . Characterization of 1.7 million TR + benchmark variants + new method to overcome var. representation issues! Great work lead by Adam E. BCM HGSC & great collab. from so many! (1/4)