Vaclav Veverka
@vaclav_veverka
scientist, structural biologist, PI (@IOCBioNMR) at @IOCBPrague and @CharlesUniPRG
ID: 174639123
https://veverka.group.uochb.cz/en 04-08-2010 12:41:48
553 Tweet
364 Followers
639 Following

We are looking for a PhD student to join our lab at IOCB Prague to study chromatin associated complexes. #structuralbiology #nmr #cryoem #transcription #epigenetics @iocbionmr uochb.cz/en/open-positi… instagram.com/p/ChsE3UfDw2C/…

My lab Caltech Chemistry & Chemical Engineering is #hiring! Looking for a postdoc to join us in sunny Pasadena and tackle questions related to IDPs, phase separation, and transcriptional control using diverse experimental approaches. Please RT. Details and how to apply: nature.com/naturecareers/… #jobs

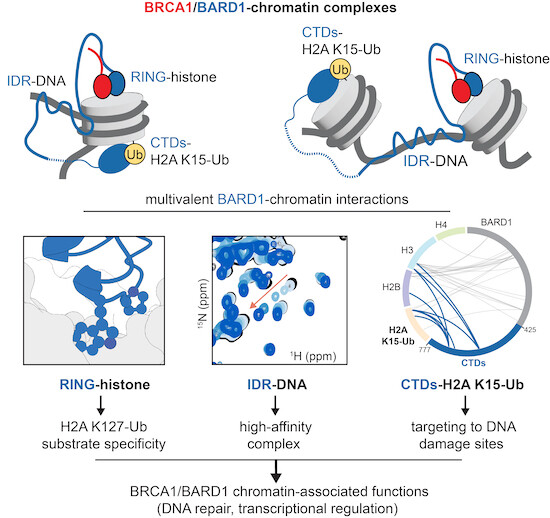
Extensive multivalent interactions between #BRCA1/#BARD1 E3 ligase IDRs & #nucleosome substrates facilitate chromatin recruitment & contribute to histone H2A ubiquitylation and to DNA repair @SamWitus et al Klevit Lab UW Biochemistry Zhao Lab UT Health San Antonio embopress.org/doi/10.15252/e…


Addicted remodeler caught red-handed. Excellent job Katerina Cermakova, cermakova.bsky.social and H. Courtney Hodges!



I am incredibly proud to present our paper on pioneer factor Sox2 that is now out in Nature Communications nature.com/articles/s4146… Work led by Sveinn Bjarnason and Jordan McIvor, with Davide Mercadante, Birthe B. Kragelund Andreas Prestel Kinga Demény, Jakob Bullerjahn.





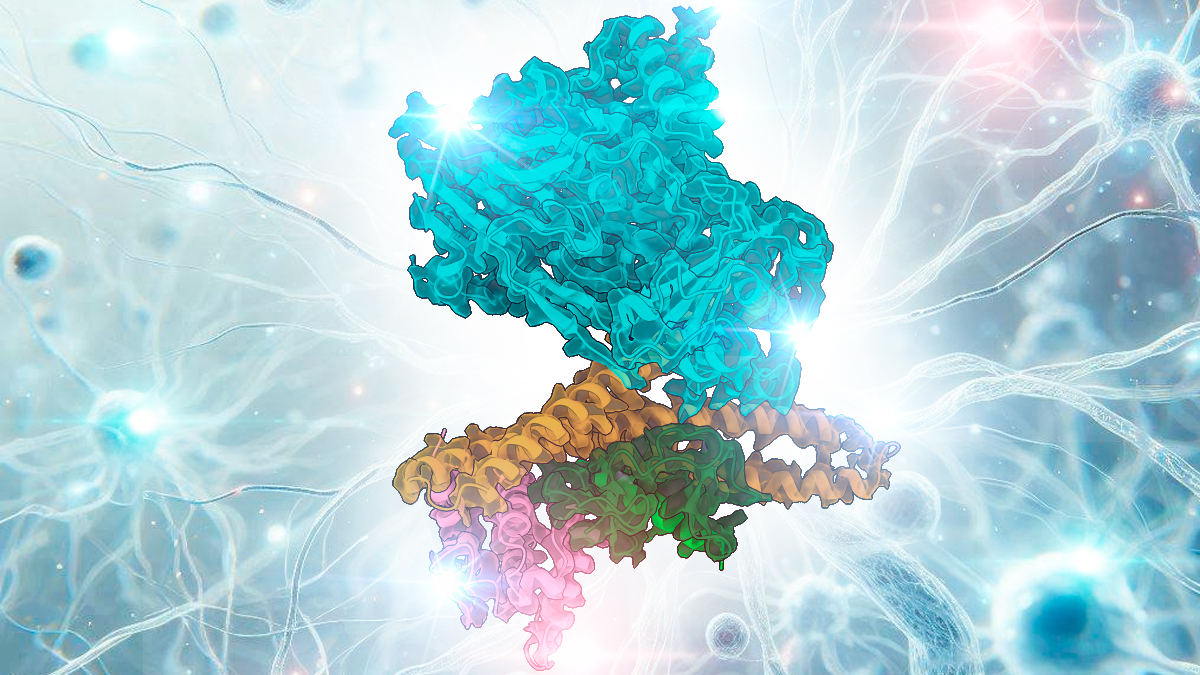
🚀 Exciting News! Our latest research on the structural basis of MICAL autoinhibition has just been published in Nature Communications ! This study sheds light on the mechanisms that control MICAL proteins, which play a critical role in cellular dynamics and processes like axon guidance.