Dr Vidya Rajasekaran-Sutherland
@vidya87_
Scientist. Colorectal cancer genetics. Equality, Diversity and Inclusion advocate. Horror aficionado. She/Her. Own views.
ID: 1097775165516247040
19-02-2019 08:29:29
44 Tweet
116 Followers
841 Following




Delighted to share the bulk of my PhD has just been published in Gut Journal! (gut.bmj.com/content/early/…). In this multidisciplinary study I co-led with Dr Vidya Rajasekaran-Sutherland, we sought to investigate how variation at a single genetic region increased colorectal cancer (CRC) risk. A 🧵: 1/










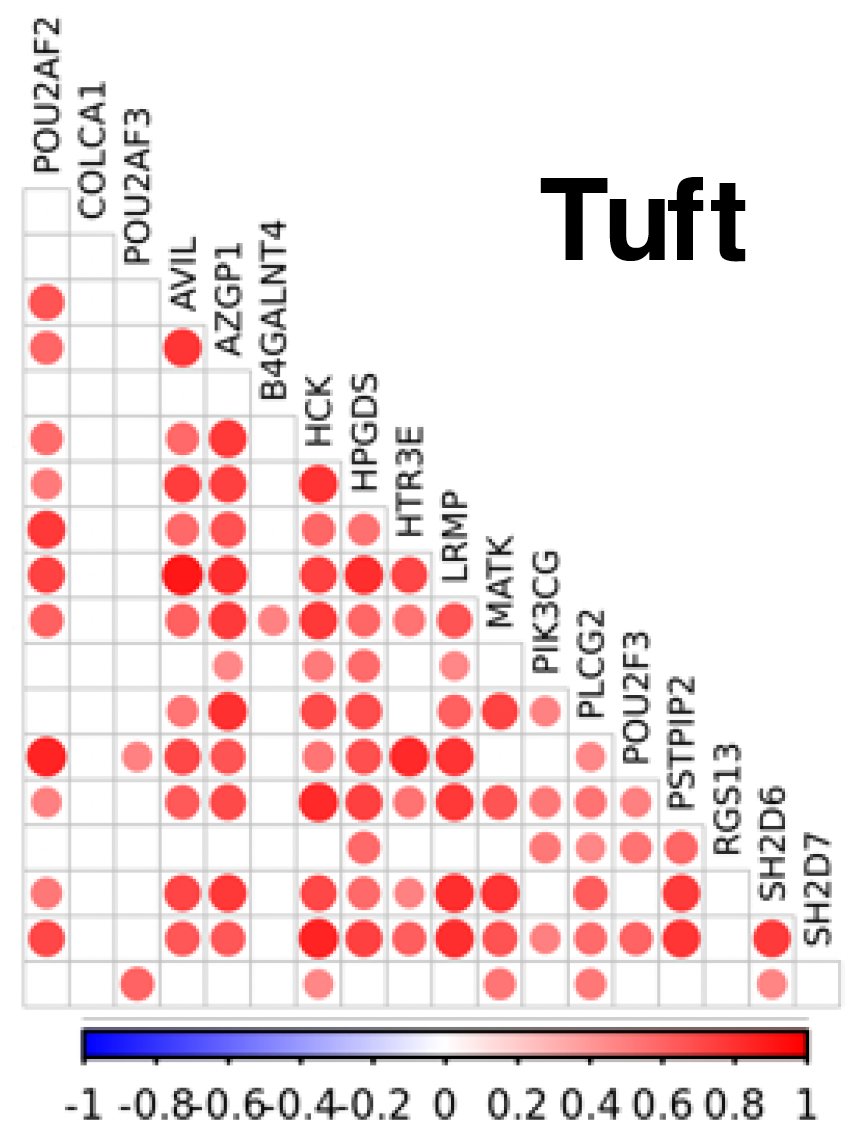
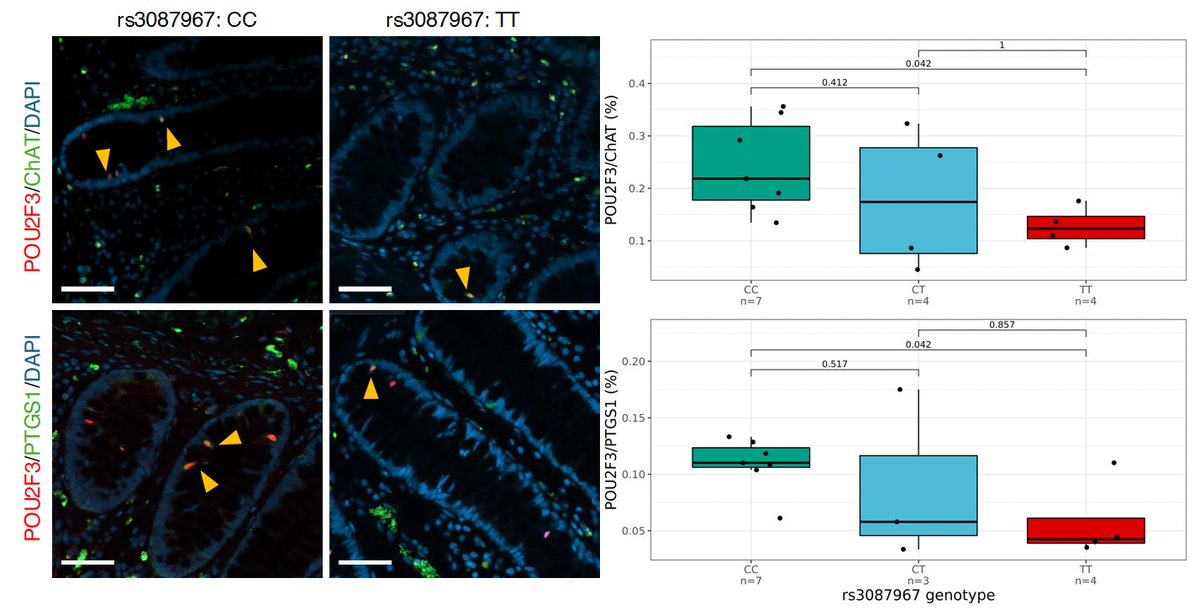
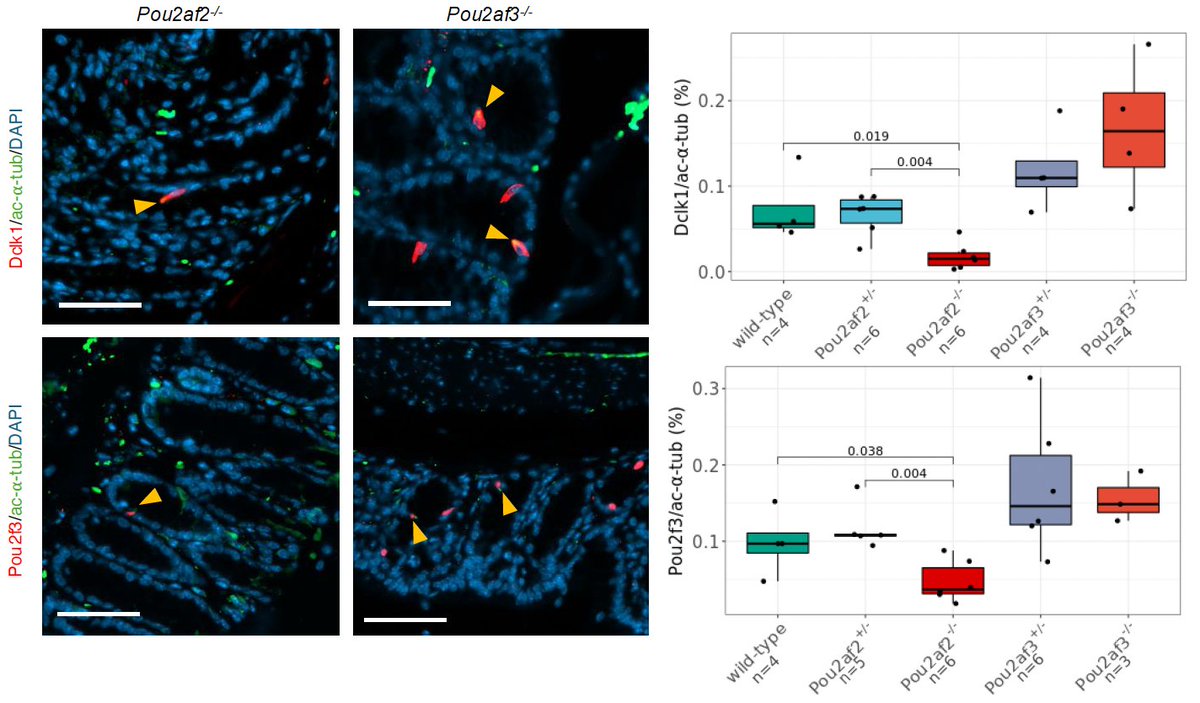
For this, we went to mice. Initiated by Dr Ruby Osborn 💙, we first validated mice as a reliable model of CRC-associated molecular phenotypes at 11q23.1, and then knocked out homologs of the cis-genes independently.
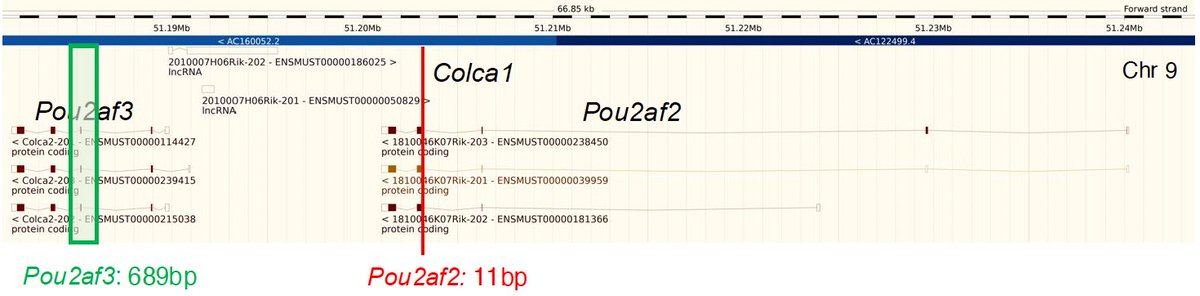





Plenty more in the paper, so do check it out! Huge thanks to all who contributed to this work: Claire Smillie, Kevin Donnelly, Marion Bacou, Edward Esiri-Bloom, Li-Yin Ooi, Morven Allan, Marion Walker, Stuart Reid, @AMeynert, Graeme Grimes, James Blackmur, Peter Vaughan-Shaw, Philip Law … 16/

Ceres Fernandez, TomlinsonGroup, Houlston Lab, ICR, Myant Lab, Farhat Din, Dr Maria Timofeeva, all greatly supervised by Malcolm Dunlop and CCGG_Edinburgh. Of course, thanks also to the donors, their families, and support from Science and Innovation at Cancer Research UK, Edinburgh Cancer Research, Institute of Genetics and Cancer!