Vikas Pejaver
@vikaspejaver
Assistant Professor @SinaiGenHealth @SinaiGenetics @IcahnMountSinai. Bioinformatics, machine learning, data science, EHR, genomic medicine. Tweets my own.
ID: 910253770767966208
19-09-2017 21:26:25
559 Tweet
332 Followers
300 Following


If interested in a postdoc position with Vikas Pejaver, send your CV, proposed research plan, and cover letter to [email protected] 🧬

My lab at Mount Sinai Institute for Genomic Health is hiring postdocs if you are interested in working with one of the most diverse EHR-linked biobanks (BioMe) and genomics of infection and immunity in diverse populations come talk to me. #ASHG2022












Join us in welcoming our new Post Doc Chad Hogan, who is joining faculty member Samira Asgari's (Samira Asgari | سمیرا عسگری) lab! #welcome #genetics #bioinformatics #infectiousdisease


Pleased to share our new preprint on using AI-generated models to explore the kinase conformational space. With Gaurav Pandey, Yan Chak (Richard) Li and others, we characterize druggable conformations via AlphaFold2, and share hundreds of these models. biorxiv.org/content/10.110…

This excellent work will be featured at the upcoming CAGI** Workshop (presentation by Clare Bycroft). Join us in Boston next week! Preliminary agenda available at genomeinterpretation.org/cagi-star-star…

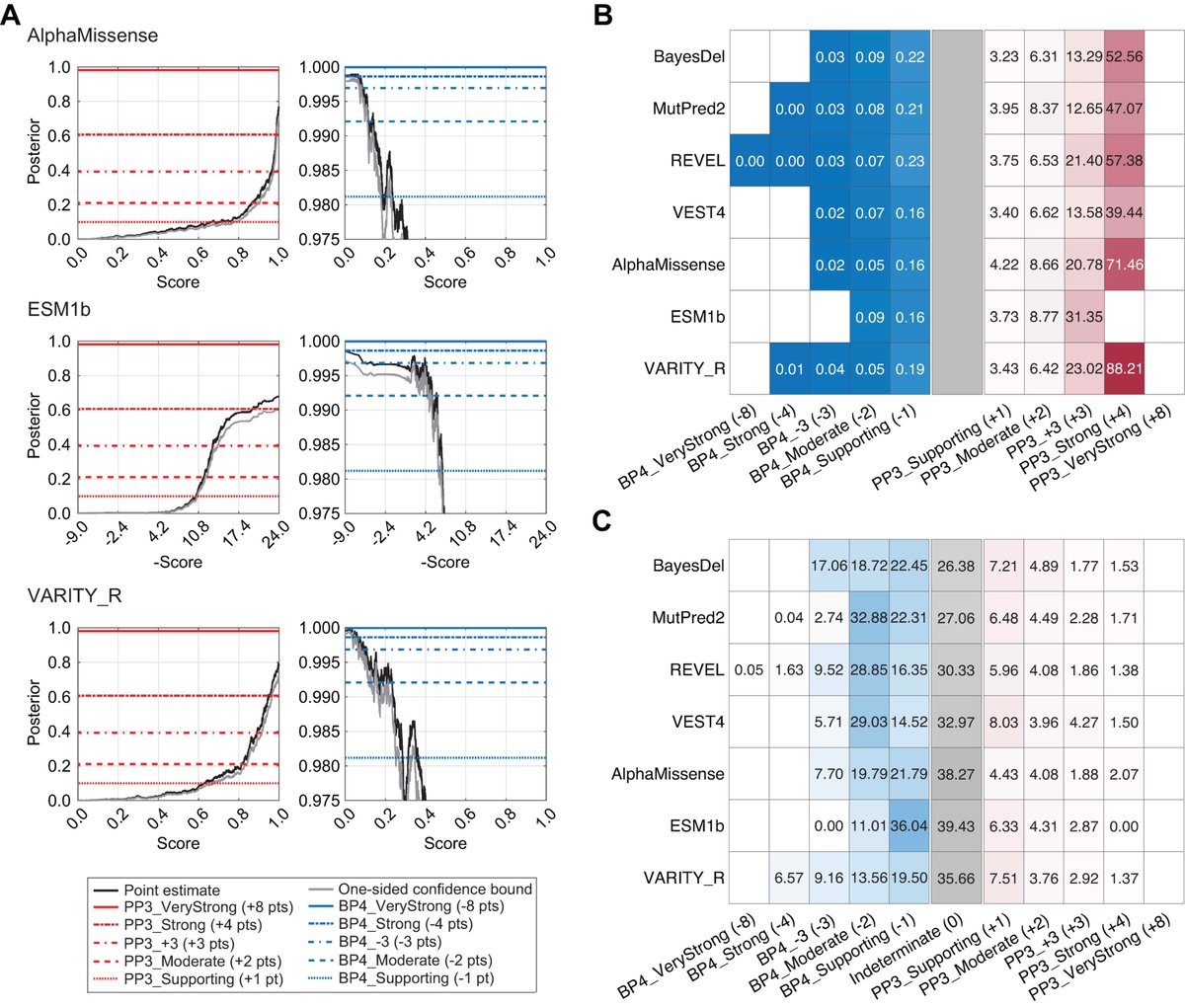
Three next-gen computational tools (AlphaMissense, ESM1b, VARITY) now calibrated for clinical variant classification! They can reach Strong evidence for pathogenicity & Moderate for benignity, performing similarly to previous tools Vikas Pejaver bit.ly/44t30N7