Shunzhi Wang
@wang_shunzhi
@BWFund CASI fellow | Postdoc @UWproteindesign | @NUChemistry @CHADNANO alum | De novo protein design for programmable self-assembly
ID: 1574449320929427456
26-09-2022 17:22:50
148 Tweet
872 Followers
673 Following




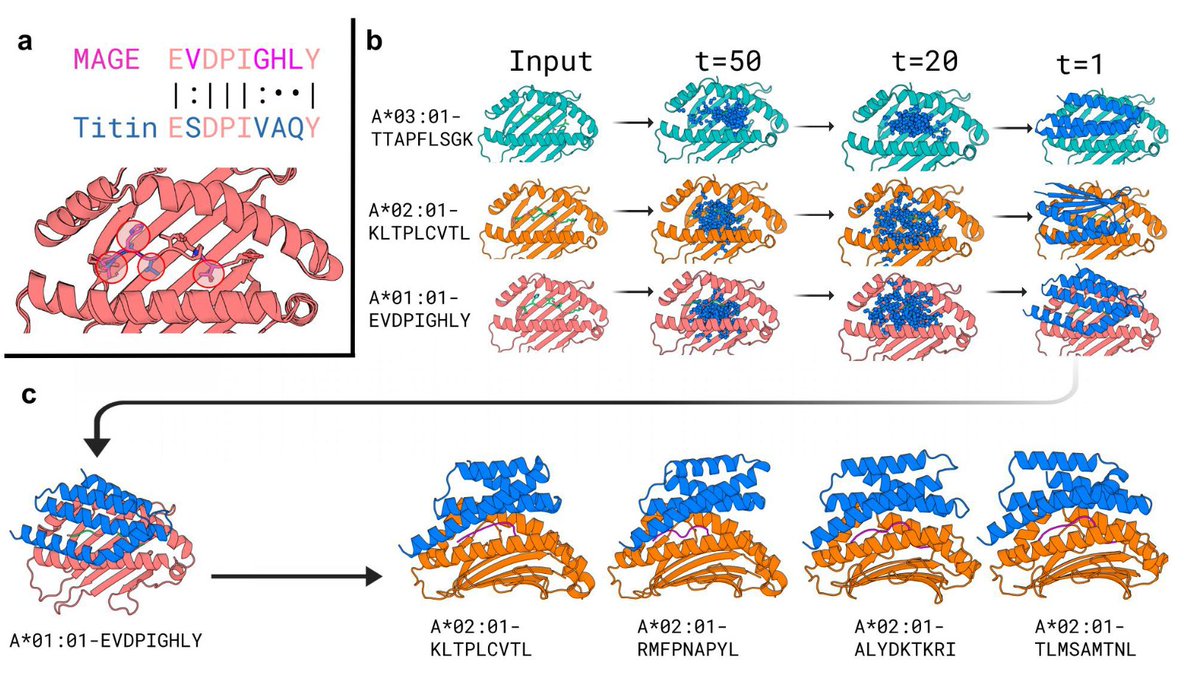
Design of high specificity binders for peptide-MHC-I complexes 🚀 New preprint from David Baker!🚀 • Researchers from Institute for Protein Design introduced a novel approach using deep learning tools like RFdiffusion to design specific protein binders for peptide-MHC-I (pMHC) complexes,


Super excited to preprint our work on developing a Biomolecular Emulator (BioEmu): Scalable emulation of protein equilibrium ensembles with generative deep learning from Microsoft Research AI for Science. #ML #AI #NeuralNetworks #Biology #AI4Science biorxiv.org/content/10.110…


In new work led by Aditi Merchant with Samuel King, we prompt engineer Evo to perform function-guided protein design with high experimental success rates, including designs that go beyond natural sequences. We also release SynGenome, the first AI-generated genomics database. 🧵 1/N






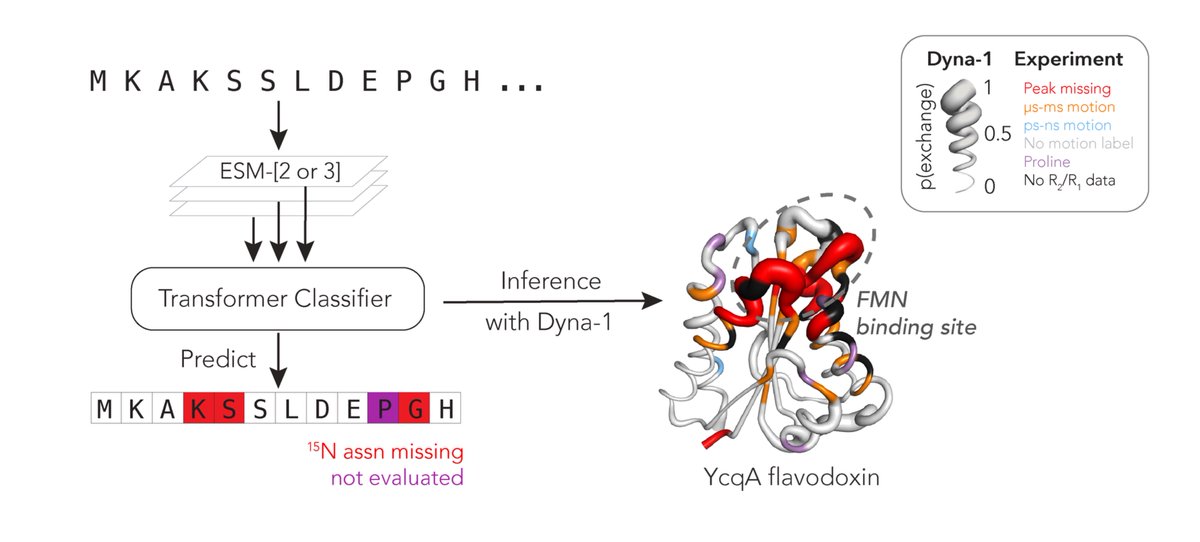
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵