WashU Epigenome Browser
@wuepgg
The WashU Epigenome Browser from @twang5 lab.
ID: 3119424783
https://epigenomegateway.wustl.edu/browser/ 26-03-2015 19:04:05
78 Tweet
385 Followers
147 Following





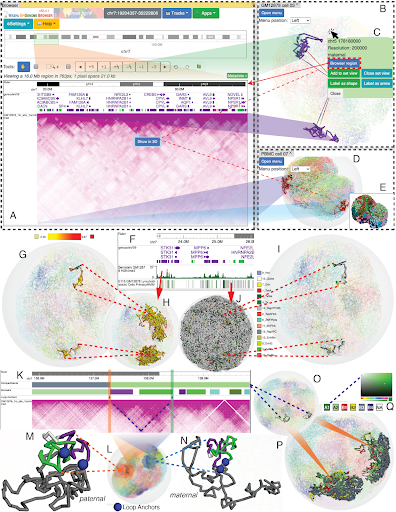
Thanks to Nature Methods for introducing our latest 3D genomics visualization expanded to the current Browser platform. Thank all our users and Wang lab Ting Wang members for suggestions and feedback! WashU Medicine Genetics


Happy to collaborated with Surag Nair from Anshul Kundaje (anshulkundaje@bluesky) lab! The dynseq browser track paper is online at Nature Genetics today! dynseq track is also integrated to higlass.io and UCSC Genome Browser. Ting Wang nature.com/articles/s4158…

Check out the Nov. in person #BrainTumorCenter seminar, featuring Ting Wang, PhD (Ting Wang) on 11/2 at 8 a.m. Dr. Wang will discuss: "Transposable elements and #cancer #epigenome evolution.” Join us! Siteman Cancer Center Barnes-Jewish Test Account McDonnell Genome Institute, WashU Medicine




Eric Roberts 39/WashU Epigenome Browser WashU Epigenome Browser. #HoffmanLabTechTalk by Coby Viner slideshare.net/hoffmanlab/was…


Joint work with @lab_lawson and Yonghao Liang on how transposable elements shape and maintain 3D genome. Thank you Nature Reviews Genetics, and apologize to those whose work we did not have space to cite. @WUSTLdbbs WashU Medicine Genetics McDonnell Genome Institute, WashU Medicine WashU Center of Reg Med nature.com/articles/s4157…





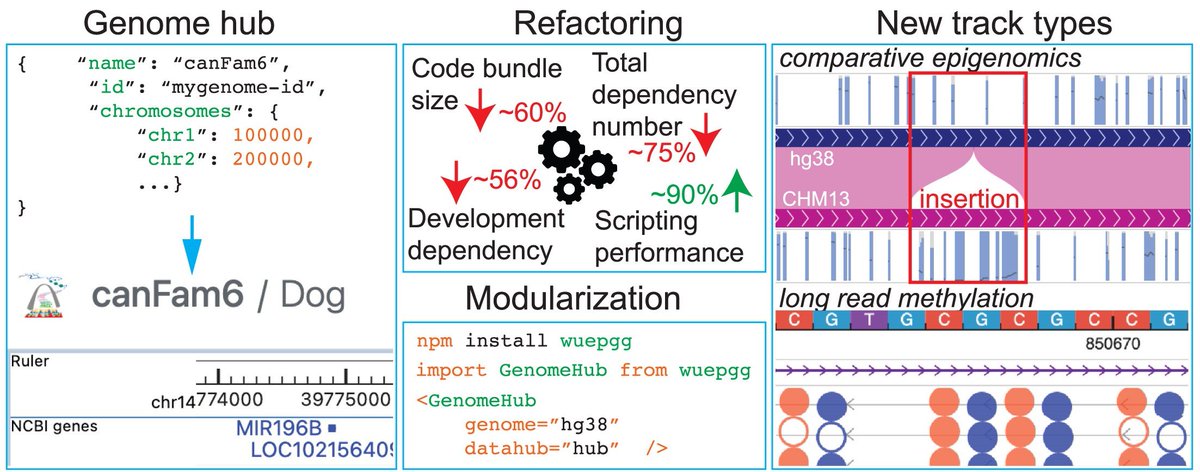
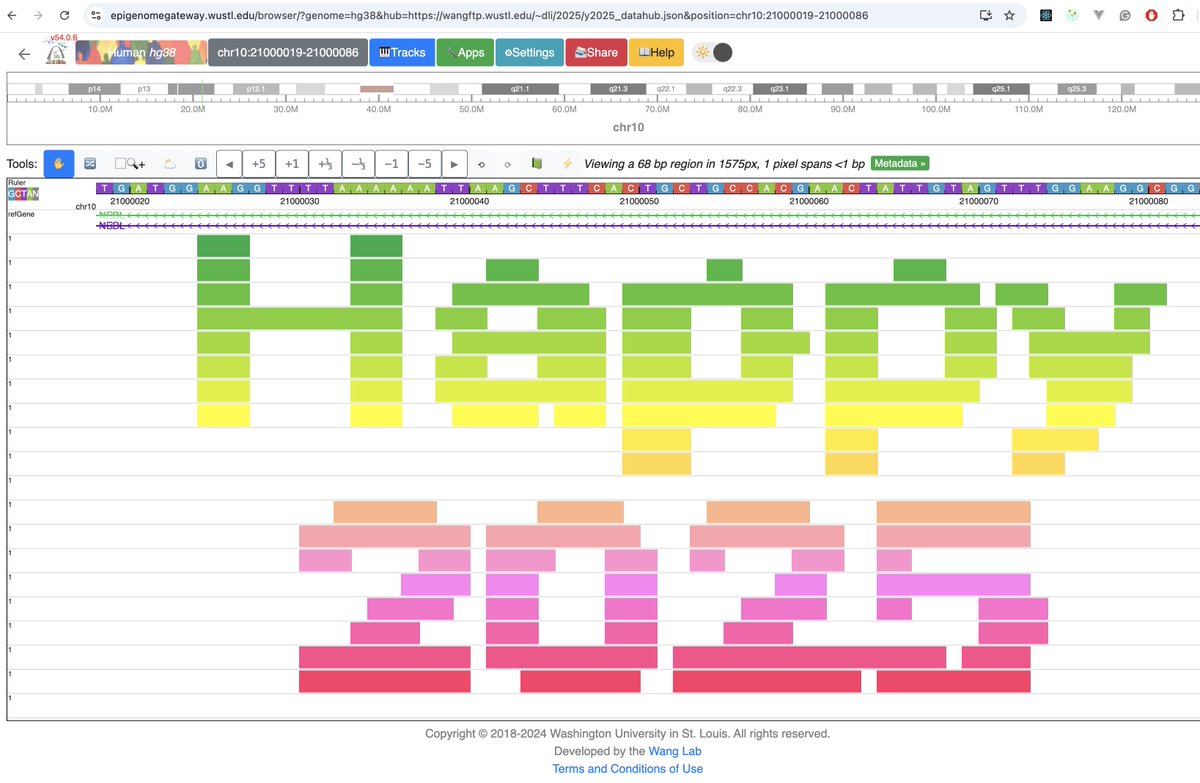
Happy to share our 2025 update manuscript now open access at Nucleic Acids Research Nucleic Acids Res. Any feedback is welcomed about the performance improvement, added features, and data. Thank you! doi.org/10.1093/nar/gk…