
Yotam Drier
@yotam_d
Genomics, Epigenomics and Topology of Cancer • Assistant Professor, Faculty of Medicine, Hebrew University of Jerusalem • yotamd.bsky.social
ID: 75747500
http://yotamdrier.ekmd.huji.ac.il/ 20-09-2009 08:19:20
398 Tweet
345 Followers
212 Following




Really awesome paper from gagneurlab on a new, clever interpretation approach showing that local DNA language models (trained on pre-defined functional elements across species) are capable of learning regulatory & structural syntax. biorxiv.org/content/10.110…

Thank you Lautenberg Center for Immunology and cancer ! and many thanks to all the students in my lab whose work made this possible.


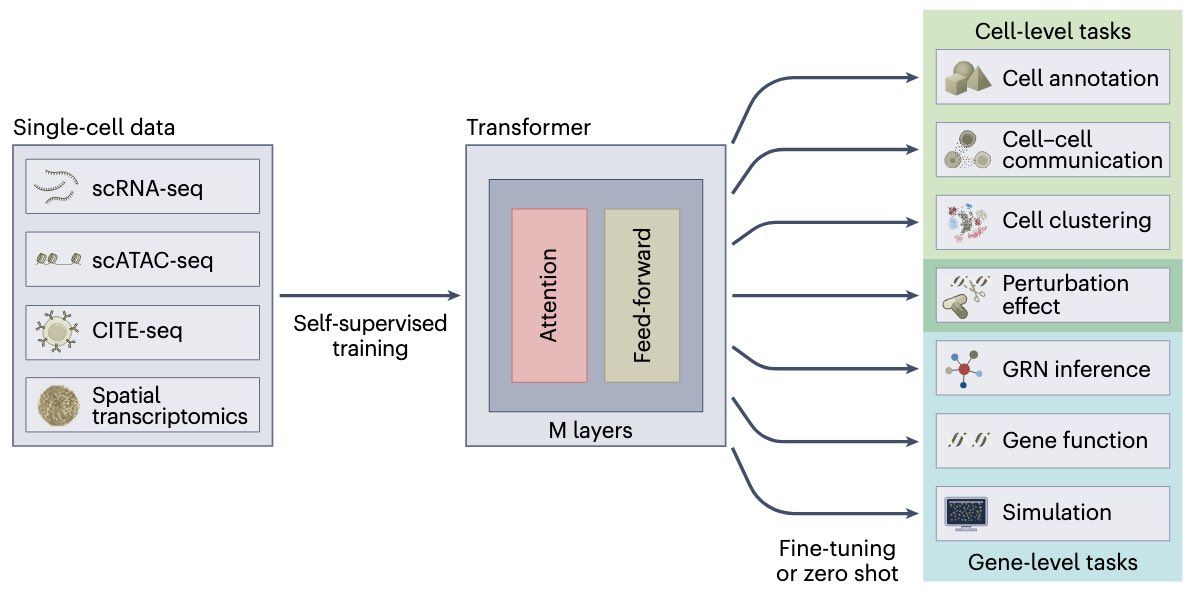
Want to learn how transformers and large-language models can help to organize&understand single cell profiles? Led by Artur Szałata, we review current such approaches in single cell genomics and give perspectives towards foundation models etc. nature.com/articles/s4159…


In this new preprint, we do a deep dive into the temporal relationship between DNA #methylation, #chromatin and #enhancers. Amazing work by first author Lindsey Guerin and thanks to our collaborators @IhrieLab, Ken Lau Lab, & biomodal. biorxiv.org/content/10.110…







Check out our review on an RNA-centric view of transcription and genome organization in an #RNA-focused issue of Molecular Cell! It was a delight to write with Richard Young. cell.com/molecular-cell…

The Vassilis Roukos lab led a new and detailed study of Topoisomerase II contribution to #3Dgenome folding in human cells and in different cell cycle phases -- my group and the MundlosLab contributed analyses/data. The paper is now published in Molecular Cell ...1/n




