Zander Bühler Harteveld
@zanderharteveld
Computational structural biology and machine learning for protein design.
ID: 1122158921450442752
27-04-2019 15:21:50
48 Tweet
219 Followers
1,1K Following

🇨🇭Swiss Secret Sauce? "Common sense and low taxes" according to The Economist. Some mention of EPFL and ETH Zürich also... economist.com/business/2022/…

Our last explorations with MaSIF and surface fingerprints for the computational design of de novo protein-protein interactions ! A lot of hard work from Pablo, @AKVanHall, Sarah, Andrea, Anthony, Michael Bronstein and collaborators biorxiv.org/content/10.110…


Using local geometries and a simple string encoding to generate existing and novel protein folds. Really happy this work w/ Zander Bühler Harteveld, Che,Yang, @SesterhennF, Stéphane and Bruno Correia is finally out! pnas.org/doi/10.1073/pn…




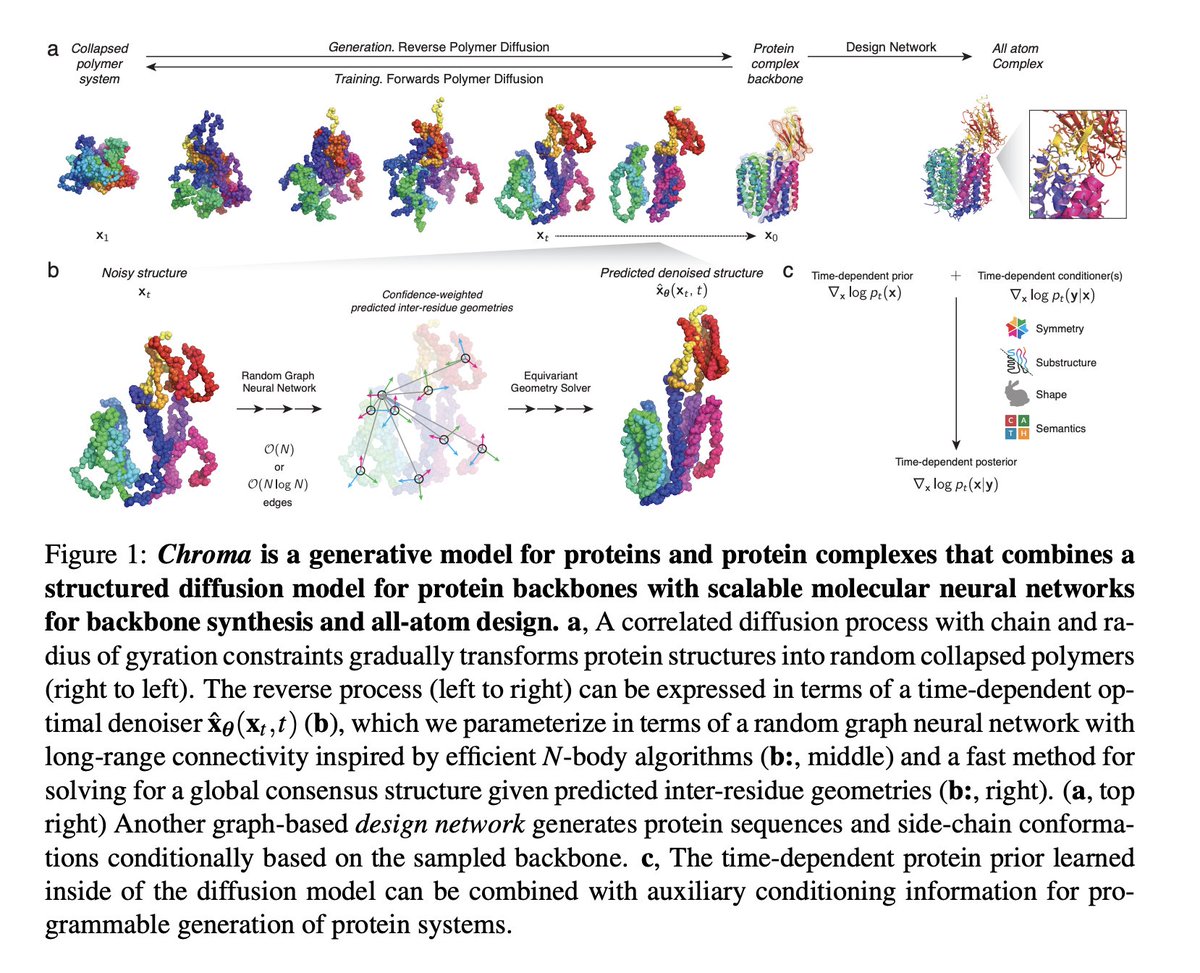
Excited to share a preprint outlining a new diffusion model for proteins named Chroma from Generate:Biomedicines! Preprint (will be on biorxiv soon): cdn.generatebiomedicines.com/assets/ingraha… Blog post: generatebiomedicines.com/chroma Some of my favorite highlights in the 🧵 below 1/N


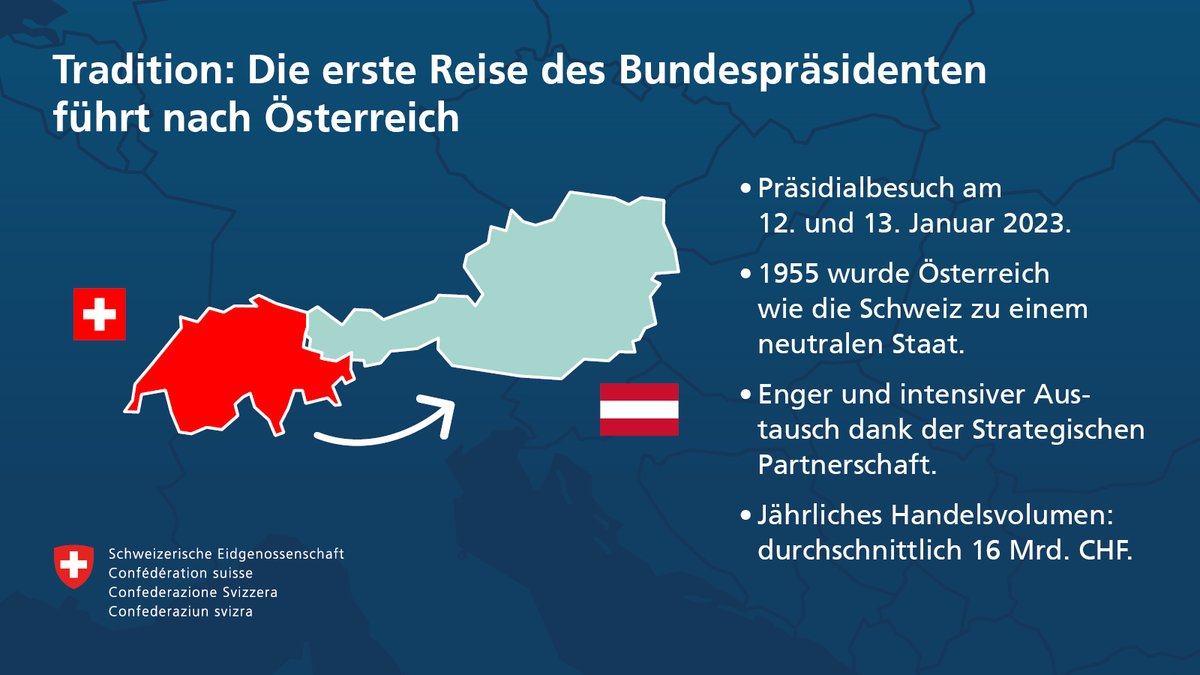
#BundespräsidentCH Alain Berset reist nach Österreich. Er wird in Wien die OSCE besuchen und mit Bundespräsident Alexander Van der Bellen und Bundeskanzler Karl Nehammer u.a. die bilateralen Beziehungen und die Sicherheitslage in Europa thematisieren. 📄admin.ch/gov/de/start/d… (BK)

Nature Biotechnology gives an overview of the use of ML & generative models for protein design, including our model Chroma. Gevorg Grigoryan shares insights about our algorithmic toolbox & how our CryoEM facility is a critical component to developing therapeutics. nature.com/articles/s4158…

Our paper with Bruno Correia Pablo Gainza-Cirauqui et al on protein design using geometric deep learning is finally in @nature We show experimental results of diverse de novo designed binders nature.com/articles/s4158…


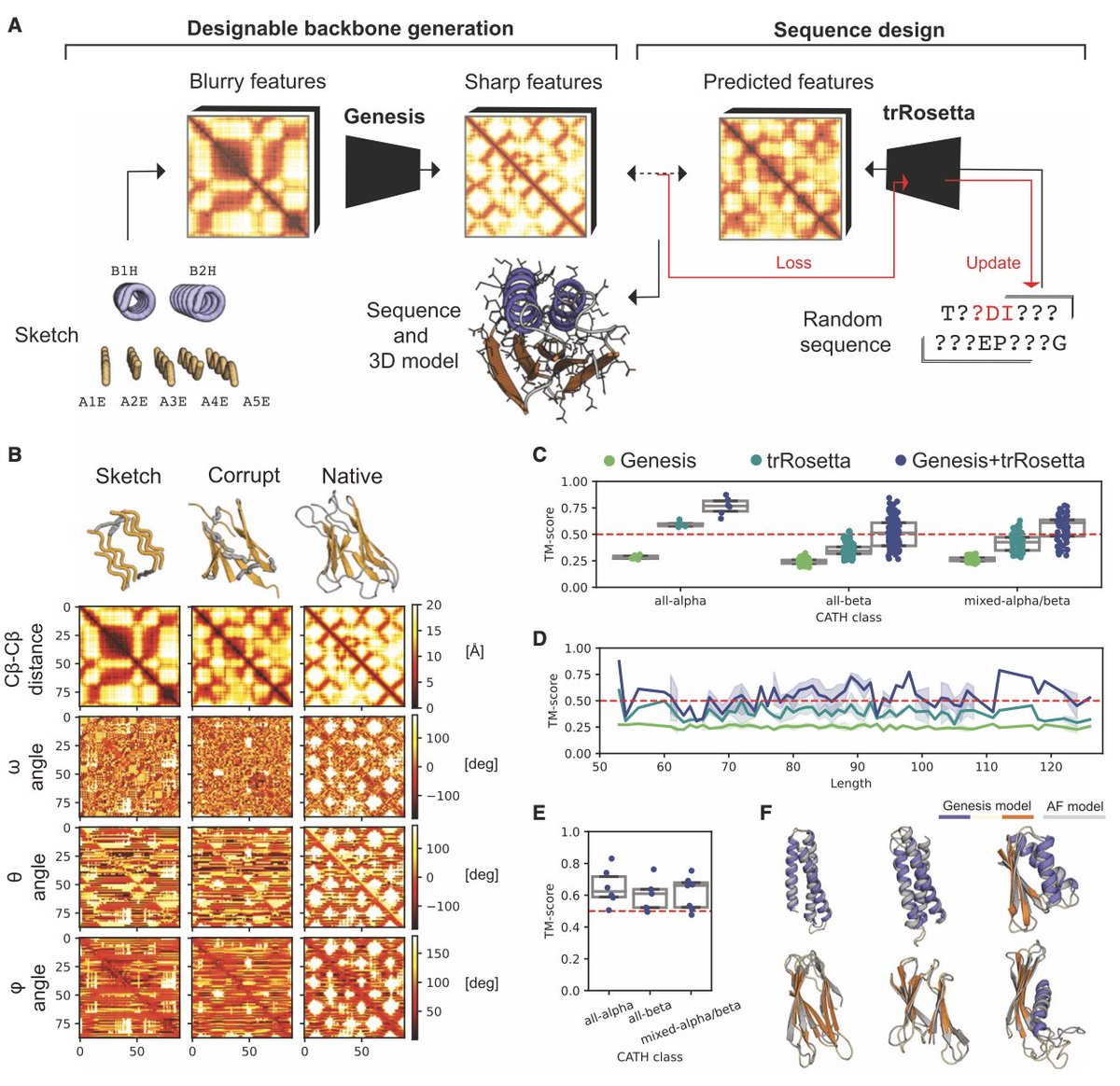
New amazing research from the Bruno Correia lab! Zander Bühler Harteveld and Alexandra explore protein topologies that have not been found in nature using a novel deep learning pipeline called Genesis


Dive into the cutting-edge world of generative AI with our CTO & Co-Founder Gevorg Grigoryan in an upcoming episode of @VantAI’s Generative AI in Drug Discovery series. Join Michael Bronstein and Bruno Correia as they gain expert perspective on the role of this technology in drug discovery.



"Exploring ‘'dark-matter' protein folds using deep learning" by EPFL , Imperial College London, Zander Bühler Harteveld, Michael Bronstein, Bruno Correia et al. "... We present a convolutional variational autoencoder that learns patterns of protein structure ..." #deeplearning #proteindesign
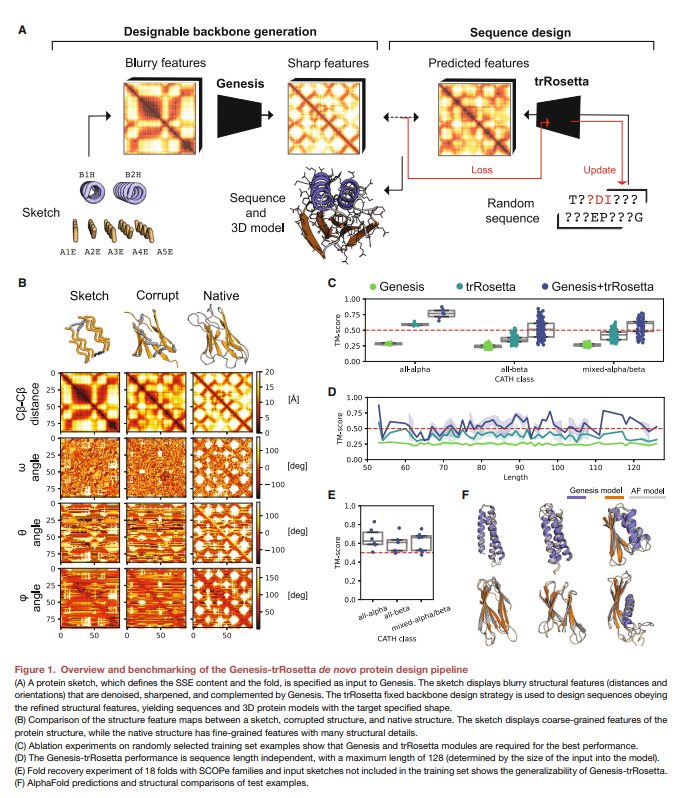

Exploring “dark-matter” protein folds using deep learning Cell Systems • Introducing Genesis VAE, a convolutional variational autoencoder that transforms low-resolution protein fold sketches into designable, stable 3D models. • Genesis VAE enables rapid exploration of