
Ziming
@zimingzzz
#DeNovoAssembly#QuantitativeTraitMapping#StressTolerence#PlantBiology#PlantPathogen
ID: 859057243635011584
01-05-2017 14:49:42
35 Tweet
53 Followers
93 Following



So great that Caroline Dean provides some intriguing unpublished data and a protein ‘Velcro’ domain……. and provides the offer for others to follow-up on this work….. #icar2022


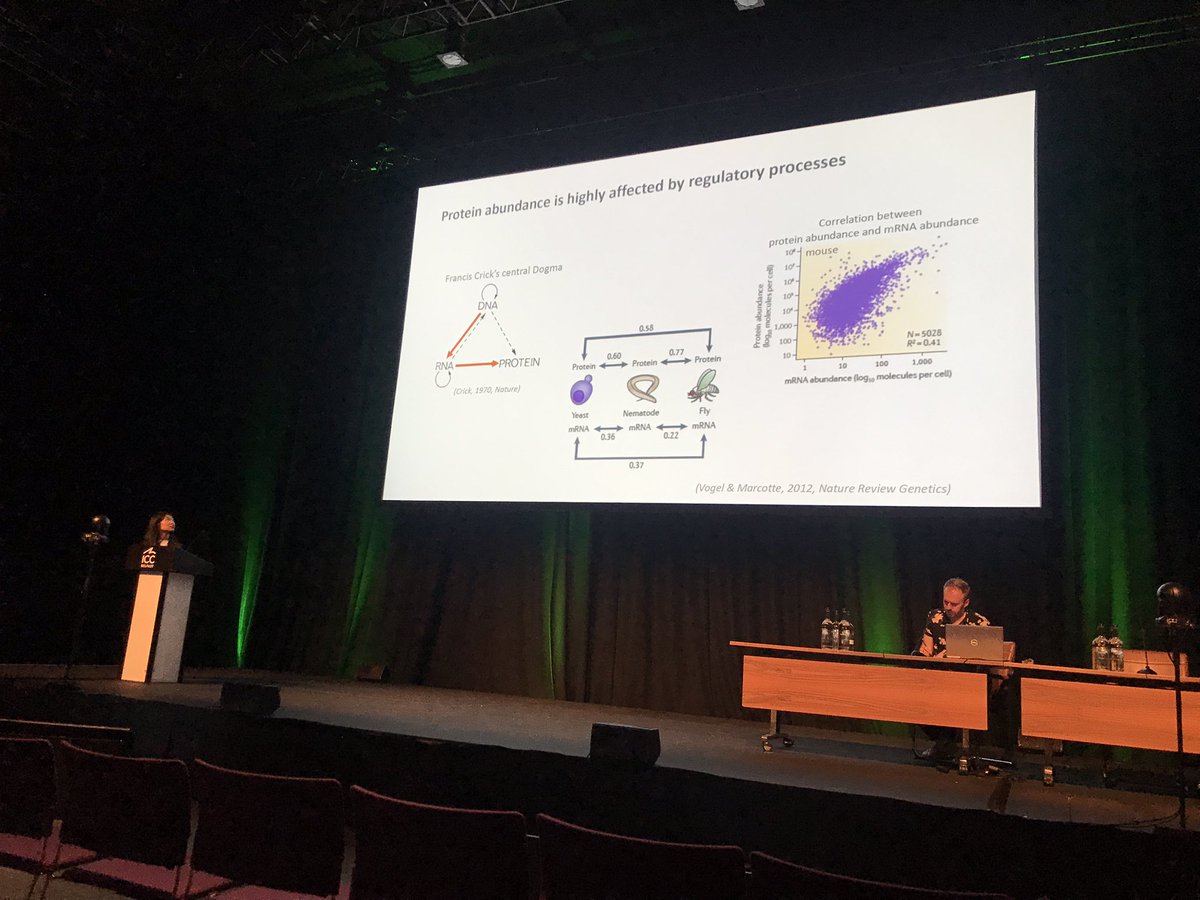
Last but not least in the #ICAR2022 dynamic proteome session, Ziming presents our collaborative project “What controls protein abundance in plants?” Richard Mott Kathryn Lilley Mark Bailey Yong-In Kim Keywan Hassani-Pak Gancho Slavov Briony J Parker

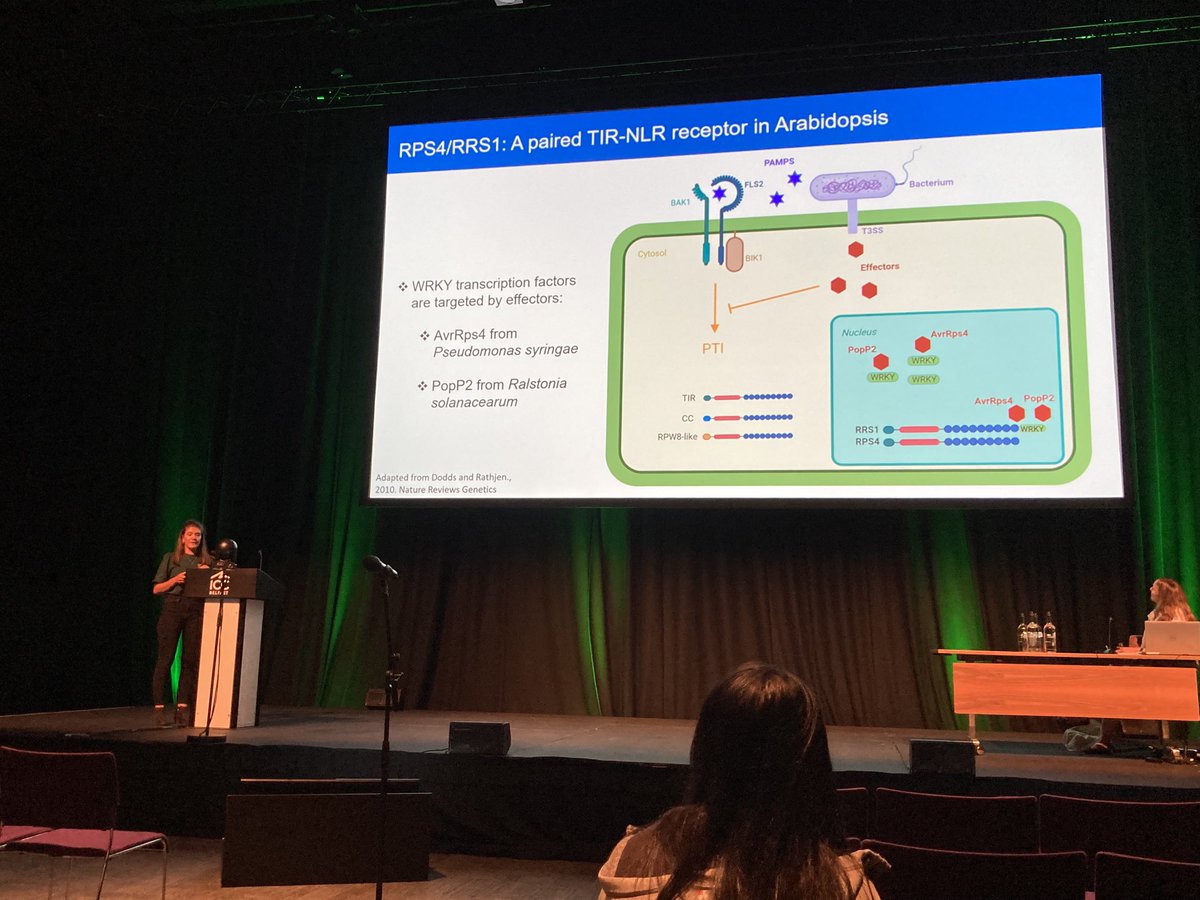
Sophie Johnson presenting her PhD project on ETI signaling using quantitative #proteomics at The Sainsbury Laboratory with Jonathan D G Jones and The Sainsbury Laboratory #proteomics team #ICAR2022



The Arabidopsis informatics session is about to start. #ICAR2022. Come if you want to hear about our new fast #GWAS implementation that enables calculation of a permutation-based significance threshold. Collaboration with Dominik Grimm Link to bioRxiv paper: biorxiv.org/content/10.110…

Nick Provart Nicholas Provart @bar-plantbio.bsky.social on the cool data visualization tools that BAR makes possible for Arabidopsis and other plants #ICAR2022





Runxuan Zhang James Hutton Institute on improving transcriptome annotation using Iso-seq data from 27 libraries of diverse samples of A. thaliana #ICAR2022





Two days left to apply for this position! We're looking for someone with #proteomics and #bioinformatics skills to join our team and work in lovely Cambridge with Kathryn Lilley #plantscijobs


We're hiring! Looking for a #bioinformatics scientist to join our Biotechnology and Biological Sciences Research #multiomics #pangenome project @rothamsted together with Kathryn Lilley Richard Mott & Keywan Hassani-Pak . We have lots of lovely #data to be analysed and scope to develop new approaches bit.ly/3ZJo2Tc








